
UDI expansion module
-
Compatible with the MERCURIUS™ product line.
-
12 UDI pairs.
-
Flexible dual indexing solutions.
Massively multiplexed library prep kits for low-input RNA - compatible with all eukaryotic cells
As low as 1 ng per well for up to 96 samples.
Compatible with Illumina® and AVITI™ sequencers.
Catalog number | #10881 |
#11881 |
#10891 |
#11891 |
|
Total preps
|
96 | 384 | 384 | 1'536 |
|
Sample multiplexing and plate format
|
96 | 96 | 384 | 384 |
|
Barcoded oligo-dT plates included
|
1 | 4 | 1 | 4 |
|
UDI pairs included
|
4 | 4 | 4 | 4 |
| Documentation | ||||
| Cat ##10881 | ||||
|---|---|---|---|---|
|
Total reactions
|
96 | |||
|
RNA multiplexing format
|
96 | |||
|
UDI pairs included
|
4 | |||
| Cat ##11881 | ||||
|---|---|---|---|---|
|
Total reactions
|
384 | |||
|
RNA multiplexing format
|
96 | |||
|
UDI pairs included
|
4 | |||
| Cat ##10891 | ||||
|---|---|---|---|---|
|
Total reactions
|
384 | |||
|
RNA multiplexing format
|
384 | |||
|
UDI pairs included
|
4 | |||
| Cat ##11891 | ||||
|---|---|---|---|---|
|
Total reactions
|
||||
|
RNA multiplexing format
|
384 | |||
|
UDI pairs included
|
4 | |||

Compatible with the MERCURIUS™ product line.
12 UDI pairs.
Flexible dual indexing solutions.

How to identify the best-performing BRB-seq method for your project.
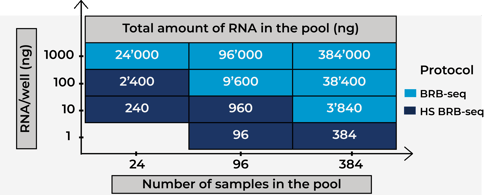
Comparison of library complexity between BRB-seq and HS BRB-seq at the same sequencing depth
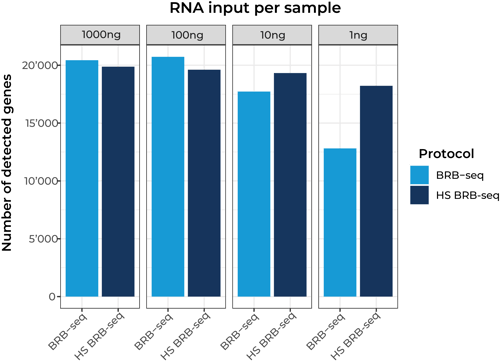
Number of detected genes between standard BRB-seq and High Sensitivity BRB-seq at different RNA inputs per sample.
96 samples were pooled together and sequenced at 2M reads/sample.
To note: all replicates were prepared from the same RNA sample.
|
For (application) |
3' mRNA sequencing |
|
For use with (equipment) |
Illumina and AVITI NGS instruments |
|
Species compatibility |
All eukaryotic species |
|
Available formats |
96 and 384 preps |
|
Shipping conditions |
Dry ice |
|
Storage conditions |
-20C |

How to identify the best-performing BRB-seq method for your project.

Comparison of library complexity between BRB-seq and HS BRB-seq at the same sequencing depth

Number of detected genes between standard BRB-seq and High Sensitivity BRB-seq at different RNA inputs per sample.
96 samples were pooled together and sequenced at 2M reads/sample.
To note: all replicates were prepared from the same RNA sample.
|
For (application) |
3' mRNA sequencing |
|
For use with (equipment) |
Illumina and AVITI NGS instruments |
|
Species compatibility |
All eukaryotic species |
|
Available formats |
96 and 384 preps |
|
Shipping conditions |
Dry ice |
|
Storage conditions |
-20C |
Each BRB-seq kit contains reagents (including four pairs of Unique Dual Indexing adapters) sufficient for the complete library preparation process for four different BRB-seq pools.
To note, the total number of RNA samples that can be processed with one kit does not exceed the kit specifications; for instance, a 96-samples kit can be used to prepare up-to 96 samples distributed across up-to four different libraries.
The recommended range of RNA amount for each sample, using the High Sensitivity BRB-seq library prep kit, is as low as 1ng per well, normally, the more RNA, the better.
The minimum recommended RIN number is 7 and the A260/230 ratio (Nanodrop) should be in the 1.5-2.2 range.
The only difference between BRB-seq and standard RNA-seq data analysis is the demultiplexing step, which is used to assign sequencing reads to their sample of origin based on the BRB-seq barcode sequence.
For a thorough description of BRB-seq data processing, please refer to the BRB-seq kit user guide.
One of the key advantages of BRB-seq is that it does not only save reagents and cost in the library preparation stage, but also in the sequencing one.
As opposed to standard RNA-seq, where 20M-30M reads per sample are required, we normally recommend to sequence BRB-seq libraries at a depth of 4M-5M reads per sample, which is normally enough to detect the vast majority of expressed genes.
The barcode set for your kit is conveniently located on the kit label. Please refer to the label for accurate identification.
For optimal compatibility, ensure that you use the appropriate plate format (e.g., for kits designed for 96 reactions, the 96 well-plate format should be used). This ensures accurate and efficient processing of your samples. If you have any further questions or concerns, please contact our support team for assistance by email or using our live chat tool.



Explore the latest, relevant publications in the industry to learn more about our technologies.
Let us do it for you. Our team will deliver the raw sequencing data (fastq files), gene count matrices and analysis report files. A cost-efficient option suitable for projects of all sizes.
Book a one-on-one call with one of our RNA experts to discover how we can assist your next project.
Receive email updates on our latest products and services, news and event updates and more. No ads, no spam.