BRB-seq technology
Unlock the power of unbiased transcriptome profiling with massively multiplexed, ultra-high-content, and high-throughput RNA sequencing.
Unlock the power of unbiased transcriptome profiling with massively multiplexed, ultra-high-content, and high-throughput RNA sequencing.



Up to 384 library preps in a single tube.
Highly optimized and rigorously evaluated sample barcodes and UMIs provide unique and sample-specific tagging of the 3' poly(A) tail of all mRNA molecules during the first step of the library preparation workflow. Samples can then be pooled and processed together in a single tube.
From purified RNA to whole blood samples.
Our solutions accommodate a wide range of inputs.
No need for prior target selection.
From purified RNA samples.
The more samples processed simultaneously, the lower the cost per sample.
MERCURIUS™ BRB-seq offers a scalable, cost-effective solution for transcriptomic analysis in large-scale blood biobank studies.
An integrated globin mRNA depletion step significantly improves transcriptome coverage and data quality from whole blood samples.
The technology excels for low-input or partially degraded RNA to unlock the full potential of archived blood samples for precision medicine, epidemiology, and disease research.

Whether for basic biology or systems-level investigations, MERCURIUS™ BRB-seq empowers researchers to push the boundaries of discovery.
By enabling transcriptome profiling across hundreds of samples simultaneously, MERCURIUS™ BRB-seq accelerates the discovery of genes linked to key agronomic traits such as yield, drought tolerance, disease resistance, and nutrient use efficiency.
This powerful tool allows researchers to decode complex plant responses to biotic and abiotic stresses, identify critical regulatory pathways, and unravel gene functions—especially in crops with large, complex genomes like wheat and maize.
MERCURIUS™ BRB-seq also supports in-depth transcriptomic studies of plant-pathogen interactions, providing insights that can drive the development of more resilient and productive crop varieties.

Distribution of the number of detected genes across 384 samples prepared with the MERCURIUS™ BRB-seq library preparation kit. The library was sequenced at an average of 1 million reads.

Number of detected genes for non-differentiated (ND) and differentiated (D) adipose stromal population cells between MERCURIUS™ BRB-seq and two other commercial kits. The expressed genes are split into three categories: Lowly Expressed (left, 0 < CPM < 10), Mid expressed (middle, 10 < CPM < 100), and Highly expressed (right, CPM > 100). To note: all replicates were prepared from the same RNA sample and the sequencing depth was 3 Million reads per sample.
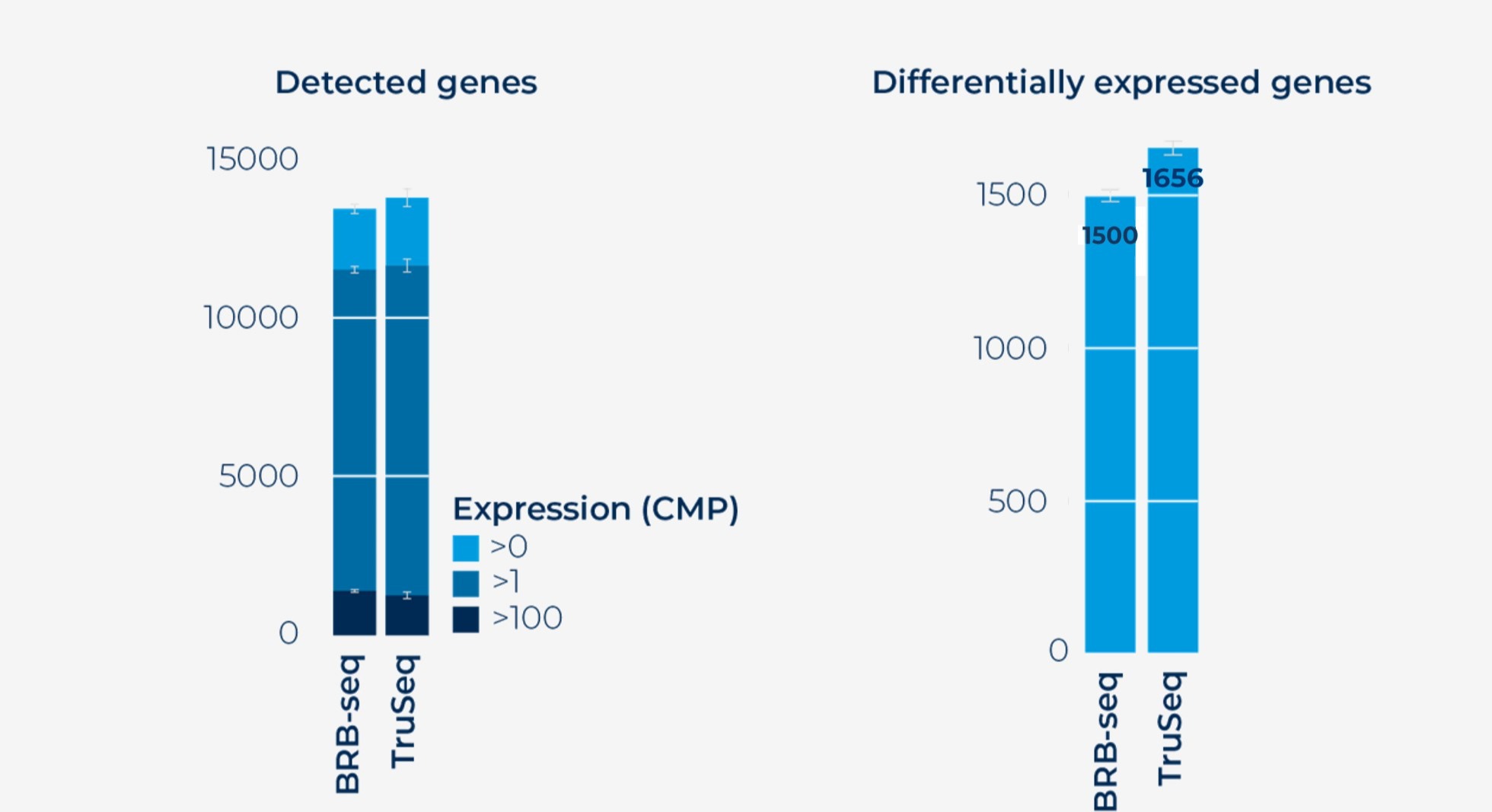
Detected genes (left) and differentially expressed genes (right) obtained on the same samples with BRB-seq and TruSeq. At comparable sequencing depth, the performance of BRB-seq data is comparable with TruSeq libraries.

The gene body coverage shows a consistent and uniform reads distribution across the entire gene body for the Full-Length BRB-seq protocol, comparable to competitor N protocol, while the BRB-seq protocol shows a significant 3' bias due to its poly-A selection methodology.
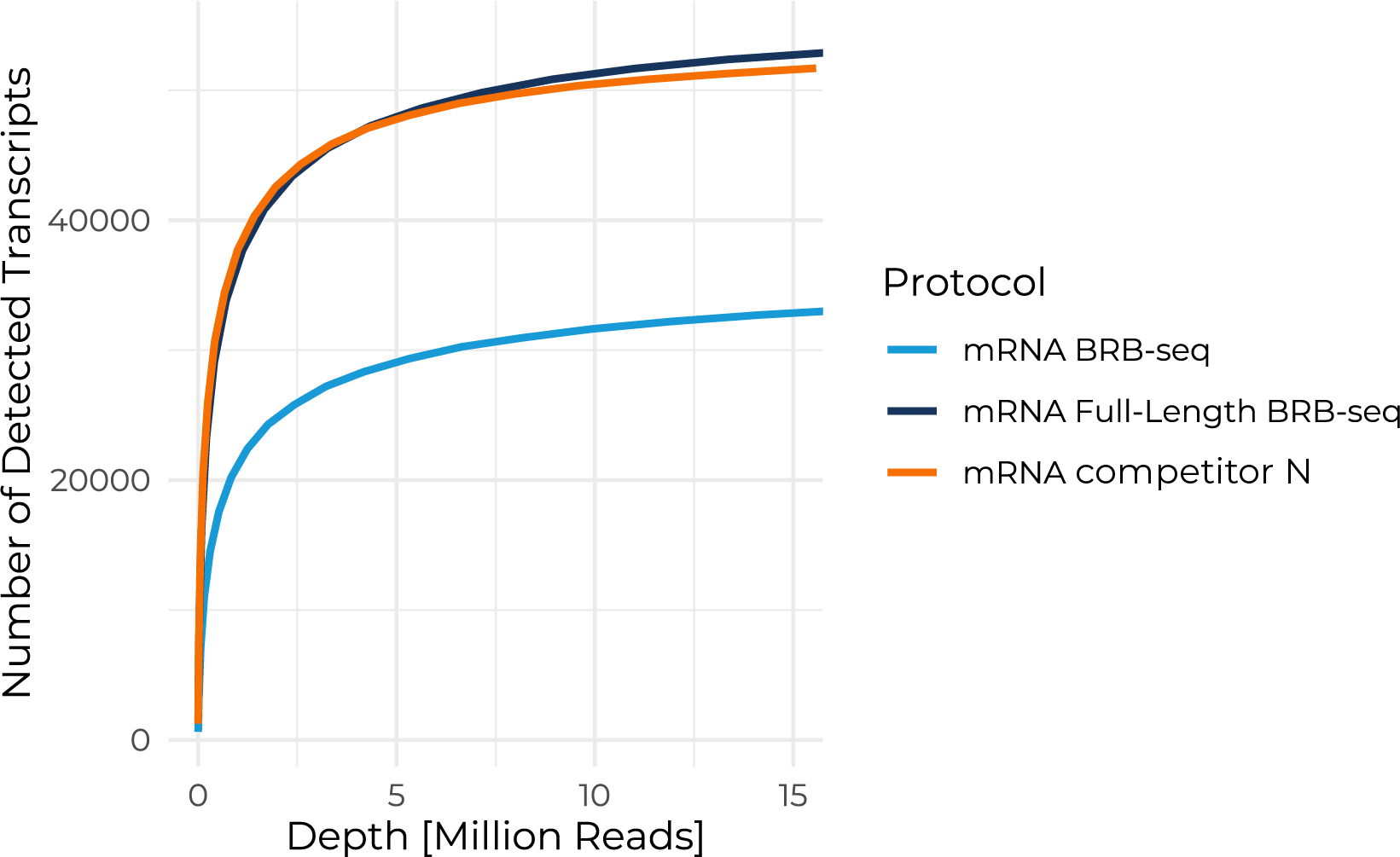
Benchmarking the Full-Length BRB-seq protocol against competitor N shows similar performance for isoform detection. More than 50,000 isoforms can be detected at a sequencing depth of 12M reads per sample.

Distribution of the number of detected genes across 48 samples prepared with the MERCURIUS™ Full-Length BRB-seq service. The library was sequenced at an average of 12 million reads.

Full-Length BRB-seq performance shows 99% demultiplexing rate from raw data, 78% mapping rate, and 18% duplication rate of the 48 pooled samples and sequenced at 12 million reads per sample.
All our kits contain all the oligos and enzymes needed to go from purified RNA samples to sequencing-ready libraries
 MERCURIUS™
MERCURIUS™
 EARLY-ACCESS
EARLY-ACCESS
MERCURIUS™
 MERCURIUS™
MERCURIUS™
 MERCURIUS™
MERCURIUS™
 MERCURIUS™
MERCURIUS™
 MERCURIUS™
MERCURIUS™
BRB-seq has proven to be an excellent method for multiplexing a large array of RNA samples for sequencing. Our team sought to generate gene expression profiles to enable our research on possible neurodegeneration therapeutic targets, requiring multiple samples for various experimental conditions. The outcomes have exceeded our expectations in terms of both result quality and cost-effectiveness, leaving us thoroughly satisfied and impressed
Dr. Emmanouil Metzakopian VP of Research and Development, bit.bio
BRB-seqwas a great way for us to multiplex large numbers of RNA samples for sequencing. We wanted a gene expression curve with a tight temporal resolution, thus, many time points for several conditions. We are very happy with the quality of the results and with the cost benefit.
Dr. Nuno Miguel Luis CNRS Researcher

Alithea Genomics Blog
21-12-2025
Identifying brain-penetrant small-molecule modulators of human microglia using a cellular model of s...

Alithea Genomics Blog
18-12-2025
DRUG-seq, first published in 2018, has transformed how pharmaceutical, cosmetics, agritech, and AI d...

Alithea Genomics Blog
18-11-2025
13 Jan 2026
Maxine Leonardi, Yves Paychère, Felix Naef. Cell-cycle inhibition preserves robust development but rebalances lineages in mouse gastruloids. bioRxiv 2026.01.08.698406
Developmental Biology
13 Jan 2026
Pham, C.N., Castelli, F., Finet, F., Leroy, C., Chollet, C., Chirayath, T.W., Moitra, S., Zarka, M., Ostertag, A., Brial, F., Combes, C., Latourte, A., Bardin, T., Fenaille, F., Richette, P. and Ea, H.K. (2026), Spermidine Reproduces the Anti-Inflammatory Effects of Intermittent Fasting and Prevents Urate and Calcium Pyrophosphate Crystal-Induced Inflammation. Arthritis Rheumatol (2025)
Inflammation
18 Nov 2025
Wu, C., Wang, T., Ghosh, A. et al. MTCH2 modulates CPT1 activity to regulate lipid metabolism of adipocytes. Nat Commun 16, 8831 (2025).
Metabolism
MERCURIUS™ BRB-seq is a transformative tool combining unbiased, ultra-high-content, and high-throughput screening with massively parallel transcriptomics.
This method uses highly optimized and rigorously evaluated sample barcodes and unique molecular identifiers to tag the 3’ poly(A) tail of all mRNA molecules in a sample-specific manner during the first-strand synthesis step of cDNA library preparation. After this step, all samples can be pooled into one single tube and processed together for the remainder of the library prep workflow.
The BRB-seq protocol is designed to work with purified RNA samples from all eukaryotic species.
MERCURIUS™ BRB-seq stands out from traditional RNA-seq methods by offering a high-throughput, cost-efficient, and streamlined workflow specifically designed for large-scale screening applications. One of the key differences lies in its sample multiplexing strategy: BRB-seq allows multiple RNA samples to be barcoded and pooled at the earliest step of the protocol—right after the RT reaction—so that the entire library preparation can proceed in a single tube. This significantly reduces both hands-on time and reagent costs.
In contrast, standard RNA-seq workflows typically require individual processing of all the RNA samples. This makes them more labor-intensive, costly, and less scalable, especially when working with large numbers of conditions, compounds, or replicates—common in drug discovery pipelines.
Each BRB-seq kit contains reagents (including four pairs of Unique Dual Indexing adapters) sufficient for the complete library preparation process for four different BRB-seq pools. To note, the total number of RNA samples that can be processed with one kit does not exceed the kit specifications; for instance, a 96-samples kit can be used to prepare up-to 96 samples distributed across up-to four different libraries.
One of the key advantages of BRB-seq is that it does not only save reagents and cost in the library preparation stage, but also in the sequencing one. As opposed to standard RNA-seq, where 20M-30M reads per sample are required, we normally recommend to sequence BRB-seq libraries at a depth of 4M-5M reads per sample, which is normally enough to detect the vast majority of expressed genes.
The only difference between BRB-seq and standard RNA-seq data analysis is the demultiplexing step, which is used to assign sequencing reads to their sample of origin based on the BRB-seq barcode sequence. For a thorough description of BRB-seq data processing, please refer to the BRB-seq kit user guide.

