Cell lysis module
-
Optimized lysis buffer for complete lysis and efficient reverse transcription.
-
Compatible with FLASH-seq library prep kits.
EARLY-ACCESS
MERCURIUS™
Ultra-sensitive, full-length and plate-based single cell RNA-seq technology for FACS-sorted cells.
Library preparation kits for Illumina®, AVITI™ and Ultima Genomics sequencers.
Catalog number | #10921 |
#10923 |
||
|
Number of samples
|
96 | 384 | ||
|
Total preps and plate format
|
96 | 384 | ||
| Documentation | ||||
Cat ##10921

|
||||
|---|---|---|---|---|
|
Total reactions
|
||||
Cat ##10923

|
||||
|---|---|---|---|---|
|
Total reactions
|
||||
Optimized lysis buffer for complete lysis and efficient reverse transcription.
Compatible with FLASH-seq library prep kits.
The MERCURIUS™ Extraction-free FLASH-seq kits contain all the oligos and enzymes needed to go from FACS-sorted cells to sequencing-ready libraries.


The number of detected genes in HEK 293T cells processed with different protocols. Reads were downsampled to 500,000 raw reads.
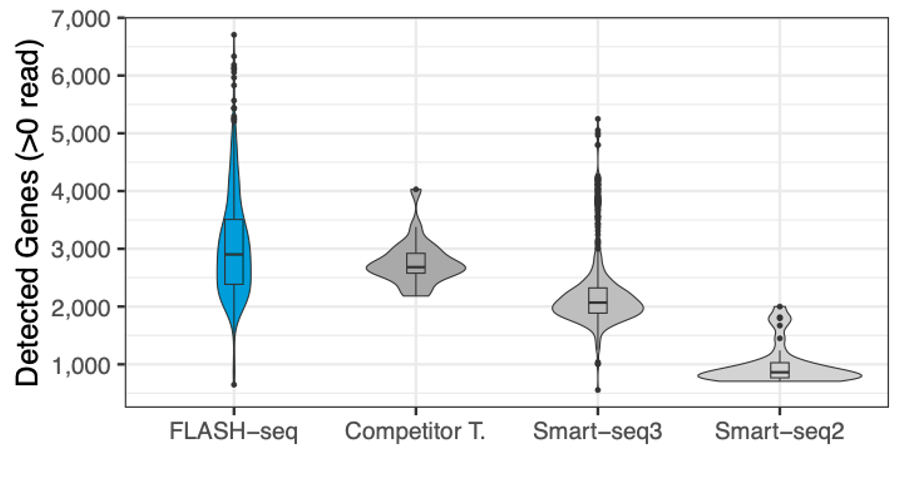
Number of genes detected in human PBMCs, processed with different protocols, and the number of reads downsampled to 125,000 raw reads.

Gene body coverage shows a uniform read distribution across the entire gene body for the FLASH-seq protocol.
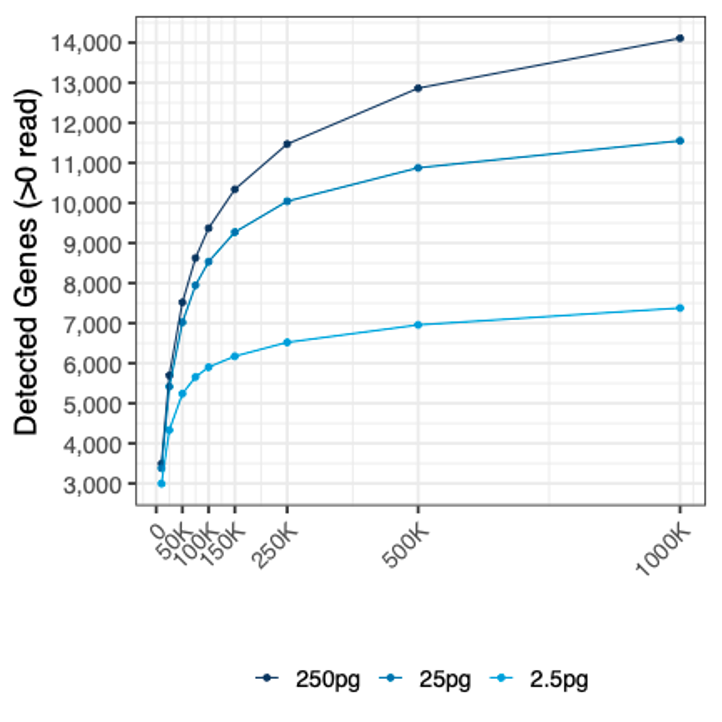
The number of genes detected in HEK 293T cells using different RNA inputs (from 2.5 pg to 250 pg) at different sequencing depths.

UMAP of hPBMC, automatically annotated using Azimuth (Hao et al., 2021).
|
For (application) |
full-length mRNA sequencing for single-cell |
|
For use with (equipment) |
Illumina, AVITI and Ultima Genomics instruments |
|
Species compatibility |
All eukaryotic species |
|
Available formats |
96 and 384 preps |
|
Shipping conditions |
Dry ice |
|
Storage conditions |
-20C |
We do not recommend very large cells, such as cardiomyocytes, or any other cell type incompatible with the FACS sorting step.


The number of detected genes in HEK 293T cells processed with different protocols. Reads were downsampled to 500,000 raw reads.

Number of genes detected in human PBMCs, processed with different protocols, and the number of reads downsampled to 125,000 raw reads.

Gene body coverage shows a uniform read distribution across the entire gene body for the FLASH-seq protocol.

The number of genes detected in HEK 293T cells using different RNA inputs (from 2.5 pg to 250 pg) at different sequencing depths.

UMAP of hPBMC, automatically annotated using Azimuth (Hao et al., 2021).
|
For (application) |
full-length mRNA sequencing for single-cell |
|
For use with (equipment) |
Illumina, AVITI and Ultima Genomics instruments |
|
Species compatibility |
All eukaryotic species |
|
Available formats |
96 and 384 preps |
|
Shipping conditions |
Dry ice |
|
Storage conditions |
-20C |
We do not recommend very large cells, such as cardiomyocytes, or any other cell type incompatible with the FACS sorting step.
We have significantly enhanced the original FLASH-seq method to offer a streamlined workflow and superior data output. This plate-based technology (available in 96- and 384-well formats) features a novel, non-toxic tagmentation buffer and delivers ultra-sensitive gene detection, capturing up to two times more genes compared to other commercially available solutions.
We do not recommend very large cells, such as the cardiomyocytes, as they are not compatible with the FACS sorting step.
For optimal results, we require the cells to be directly FACS-sorted in the dedicated well plates. The 96- and 384-plates contain the lysis buffer. It is important for the user to sort the cells in the middle of the well. Please refer to the User Guide and the Sample Submission Guidelines for more details.
The average recommended sequencing depth is 250'000 reads/cell.



Explore the latest, relevant publications in the industry to learn more about our technologies.
Let us do it for you. Our team will deliver the raw sequencing data (fastq files), gene count matrices and analysis report files. A cost-efficient option suitable for projects of all sizes.
Book a one-on-one call with one of our RNA experts to discover how we can assist your next project.
Receive email updates on our latest products and services, news and event updates and more. No ads, no spam.