DRUG-seq technology
Extraction-free and high-throughput RNA-seq technology designed to provide unbiased and high-content full transcriptome profiling across thousands of samples in parallel at low cost and short turnaround times.
Extraction-free and high-throughput RNA-seq technology designed to provide unbiased and high-content full transcriptome profiling across thousands of samples in parallel at low cost and short turnaround times.



MERCURIUS™ DRUG-seq is a powerful solution for compound screening and drug discovery, enabling ultra-high-content, unbiased, and high-throughput profiling with extraction-free transcriptomics. The method leverages rigorously optimized sample barcodes and unique molecular identifiers (UMIs) to label the 3' poly(A) tails of mRNA molecules during the first-strand cDNA synthesis. This efficient tagging approach ensures precise sample identification, robust transcript quantification, and seamless scalability for large screening campaigns.
Skip tedious RNA extraction steps and go straight to library prep.
Up-to 384 samples processed in one single tube!
Our highly optimized sets of barcoded primers uniquely "tag" individual RNA samples during the first step of library preparation so that you can pool and process all samples together in a single tube early in the workflow.
Improved protocol for higher mapping and gene detection rates
MERCURIUS™ DRUG-seq leverages a high-yield enzyme for reverse transcription. This streamlines the workflow and eliminates potential biases and duplication introduced with pre-amplification and template-switching oligos (TSO) used in the original DRUG-seq protocol while delivering superior performance.
Massively multiplexed workflow with 96 and 384-well plate formats so that you can screen thousands of samples in parallel at scale.
No need for prior target selection.
No pre-amplification needed, leading to higher mapping and gene detection rates.
The more samples processed simultaneously, the lower the cost per sample.
Enables seamless integration into high-throughput workflows, further reducing hands-on time and increasing scalability, reproducibility and efficiency.
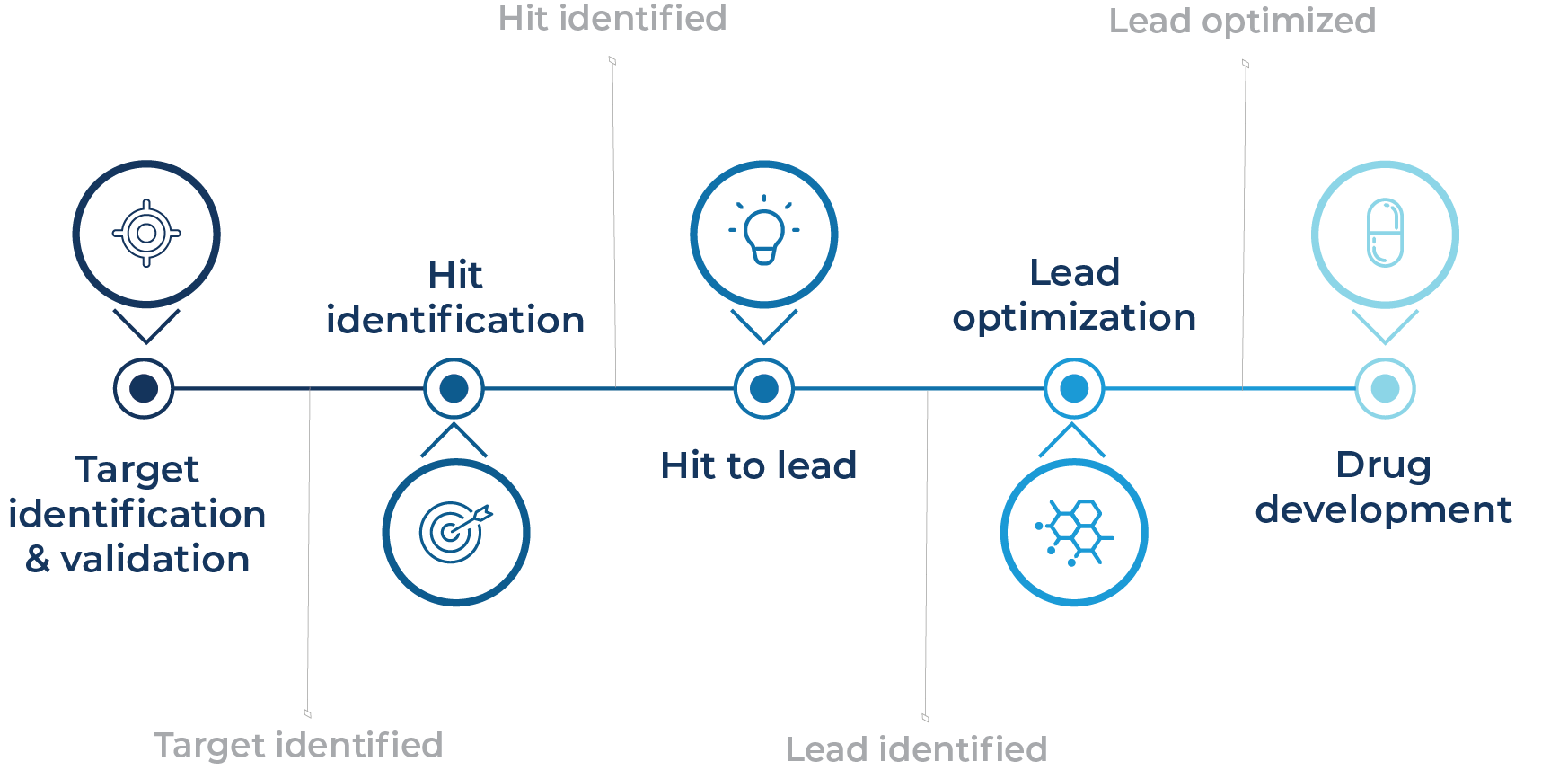
A scalable and cost-effective transcriptomic profiling solution that accelerates target identification and validation, hit identification, hit-to-lead, and lead optimization.
By providing deep gene expression insights across thousands of compounds, MERCURIUS™ DRUG-seq reveals mechanisms of action, detects on/off-target effects, and uncovers toxicity markers, empowering faster, data-driven decisions throughout the drug development pipeline.

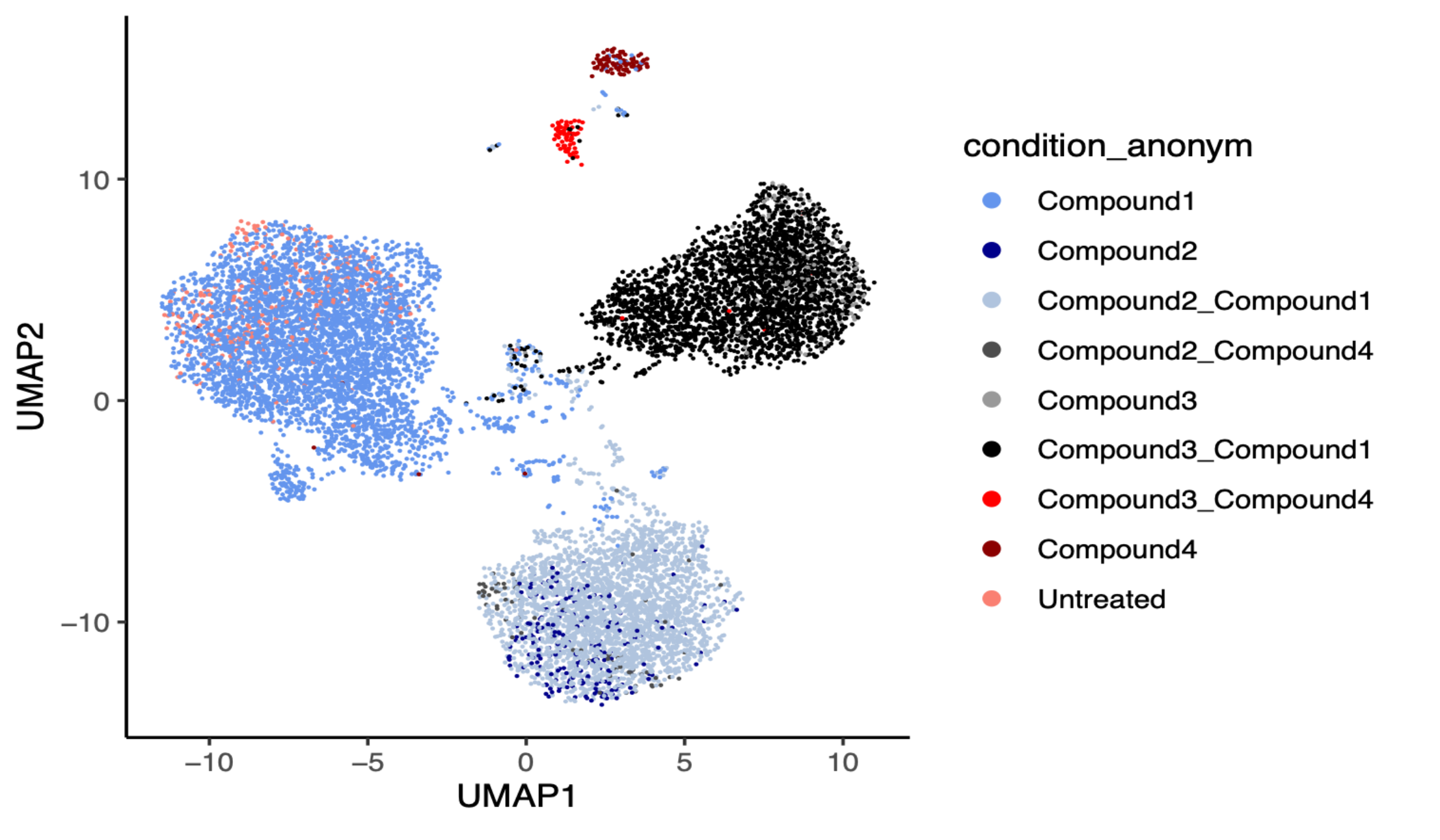
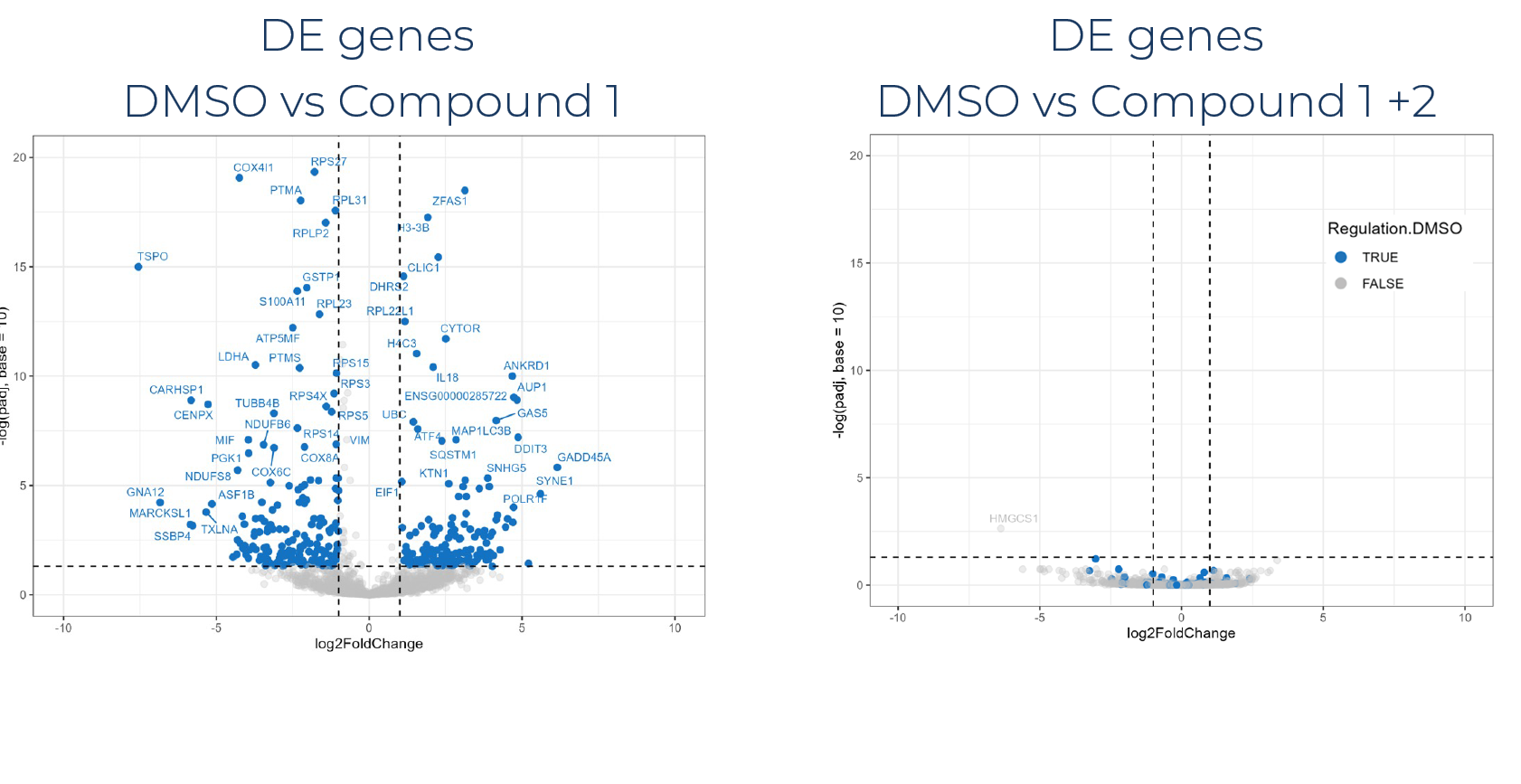
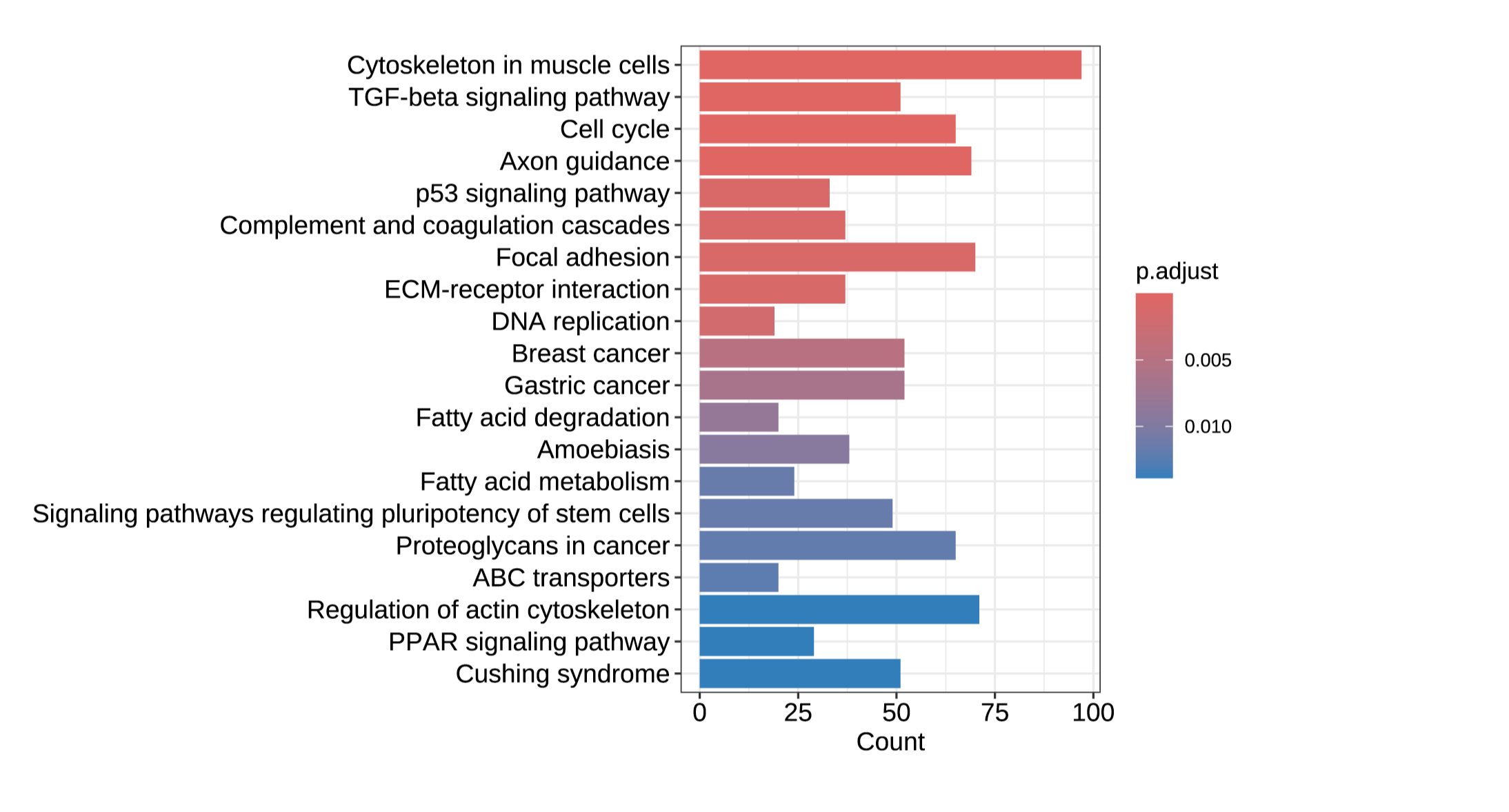
Artificial intelligence algorithms require vast quantities of robust, in-depth training data for pattern recognition and predictive modeling before researchers can extract meaningful insights with confidence. The unparalleled scalability and cost-effectiveness of MERCURIUS™ DRUG-seq now enables researchers to generate high-throughput transcriptome-wide data for thousands of samples or conditions. These data help to train advanced machine learning models and deep learning algorithms, streamlining screening pipelines and enabling smarter, faster compound discovery.
This integration facilitates scalable phenotypic screening and compound similarity mapping, accelerating mechanism of action identification, target deconvolution, and gene network analysis. Leveraging AI-powered pattern recognition and predictive modeling, researchers can extract meaningful gene signatures and uncover novel biomarkers with greater confidence.
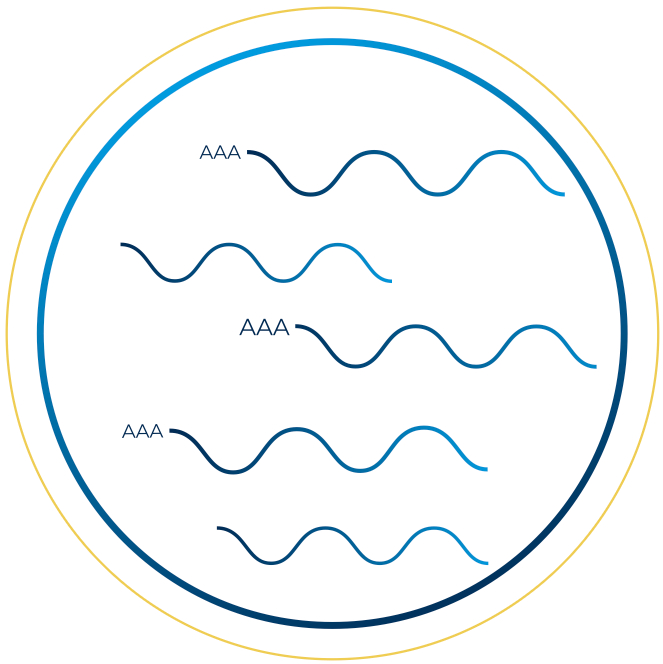
Capture the complete transcriptome by sequencing all RNA molecules within a sample, covering their full length from the 5′ to 3′ ends.
This approach enables a comprehensive analysis of transcript isoforms, alternative splicing events, non-coding RNAs, and overall gene expression profiles.
All our kits contain all the oligos and enzymes needed to go from 2D cell cultures to sequencing-ready libraries.
MERCURIUS™


EARLY-ACCESS
MERCURIUS™
MERCURIUS™

Choose from library prep only or complete end-to-end services, including cell culture and treatment, Cell Painting, NGS, and downstream analysis.
The DRUG-seq platform enabled us to systematically profile compound-induced transcriptional responses at scale, generating a rich dataset that helped prioritize testable hypotheses. The team at Alithea provided outstanding technical guidance throughout, including support in developing a customized analysis pipeline tailored to our needs. The turnaround time was fast, the process seamless, and the collaboration highly productive. We would be glad to work together again.
Mikołaj Słabicki, Ph.D. Principal Investigator at the MGH Krantz Family Center for Cancer Research. Assistant Professor of Medicine at Harvard Medical School. Affiliate Faculty Member at the Broad Institute of MIT and Harvard
DRUG-seq has completely transformed how I approach and think about transcriptomic screening. It's fast, scalable, and incredibly cost-effective—perfect for profiling hundreds of compounds in parallel. In my experience, it’s one of the most powerful tools for uncovering mechanisms of action and capturing transcriptional signatures at scale. I've used DRUG-seq to profile different cells treated under time points, dose responses, combination treatments, and a variety of complex cellular models, from cardiomyocytes to organoids—and it consistently delivers high-quality, actionable data. The ability to pool samples early without compromising data quality truly makes it a game-changer.
Andrea Hadjikyriacou, Ph.D. Principal Scientist I, Novartis Biomedical Research

Alithea Genomics Blog
24-02-2026
DRUG-seq is a cost-effective, high-throughput bulk RNA-seq library preparation method for compound s...

Alithea Genomics Blog
21-12-2025
Identifying brain-penetrant small-molecule modulators of human microglia using a cellular model of s...

Alithea Genomics Blog
18-12-2025
DRUG-seq, first published in 2018, has transformed how pharmaceutical, cosmetics, agritech, and AI d...
13 Jan 2026
Maxine Leonardi, Yves Paychère, Felix Naef. Cell-cycle inhibition preserves robust development but rebalances lineages in mouse gastruloids. bioRxiv 2026.01.08.698406
Developmental Biology
13 Jan 2026
Pham, C.N., Castelli, F., Finet, F., Leroy, C., Chollet, C., Chirayath, T.W., Moitra, S., Zarka, M., Ostertag, A., Brial, F., Combes, C., Latourte, A., Bardin, T., Fenaille, F., Richette, P. and Ea, H.K. (2026), Spermidine Reproduces the Anti-Inflammatory Effects of Intermittent Fasting and Prevents Urate and Calcium Pyrophosphate Crystal-Induced Inflammation. Arthritis Rheumatol (2025)
Inflammation
18 Nov 2025
Wu, C., Wang, T., Ghosh, A. et al. MTCH2 modulates CPT1 activity to regulate lipid metabolism of adipocytes. Nat Commun 16, 8831 (2025).
Metabolism
MERCURIUS™ DRUG-seq is a transformative tool for compound screening and drug discovery, combining unbiased, ultra-high-content, and high-throughput compound screening with massively parallel and extraction-free transcriptomics. This metho d uses highly optimized and rigorously evaluated sample barcodes and unique molecular identifiers to tag the 3’ poly(A) tail of all mRNA molecules in a sample-specific manner during the first-strand synthesis step of cDNA library preparation.
The DRUG-seq protocol is designed to work with frozen cells and bypass RNA extraction. Thanks to the highly optimized lysis buffers for cell lysis of 2D cell cultures and organoid models, it efficiently generates library preps without prior RNA isolation.
MERCURIUS™ DRUG-seq stands out from traditional RNA-seq methods by offering a high-throughput, cost-efficient, and streamlined workflow specifically designed for large-scale screening applications. One of the key differences lies in its sample multiplexing strategy: DRUG-seq allows multiple RNA samples to be barcoded and pooled at the earliest step of the protocol—right after cell lysis—so that the entire library preparation can proceed in a single tube. This significantly reduces both hands-on time and reagent costs.
In contrast, standard RNA-seq workflows typically require individual RNA extraction and processing for each sample, followed by separate library preparations. This makes them more labor-intensive, costly, and less scalable, especially when working with large numbers of conditions, compounds, or replicates—common in drug discovery pipelines.
MERCURIUS™ DRUG-seq is used for:
Drug discovery: Identifying new drug candidates and understanding how existing drugs work at a molecular level.
Mechanism of action studies: Understanding how drugs affect cellular pathways and gene regulation.
Drug repurposing: Identifying new uses for existing drugs based on gene expression changes.
Personalized medicine: Tailoring treatments based on individual genetic responses to drugs.
MERCURIUS™ DRUG-seq can be performed on various sample types, including: Cell lines, Primary cells, Organoids or Spheroids.
Validated cells lines
| Cell line/Tissue | Organism |
| HT1080 SMARCA4 KO | Human |
| hTEC | Human |
| iPSC microglia | Human |
| MCF7 | Human |
| HepG2_IGF1RKO | Human |
| Beas-2B | Human |
| dTHP-1 | Human |
| iPSC derived cardiomyocytes | Human |
| Endothelial | Human |
| Hepatocyte | Human |
| HEPG2 | Human |
| Hepatocyte | Human |
| SF9 | Spodoptera |
| A549 | Human |
| Breast Cancer (MCF7) | Human |
| B lymphoblast MM.1s. | Human |
| Hek293 | Human |
| HeLa Cells | Human |
| U2-OS | Human |
| Hek293 | Human |
| dTHP-1 | Human |
| AsPC-1 | Human |
| PBMC (TCD4) | Human |
| Skeletal muscle (LHCN-M2) | Human |
| HepaRG | Human |
| Macrophage (MV-4-11) | Human |
| PBMCs (Blood) | Human |
| Microglia | Human |
| Fibroblast Like Synoviocyte donor 1502 | Human |
| Fibroblast | Human |
| iPSC-derived neuron | Human |
| Human lung carcinoma epithelial cell line | Human |
| Human breast epithelial adenocarcinoma cell line | Human |
| Human liver hepatocellular carcinoma cell line | Human |
| Human cardiomyocytes derived from induced pluripotent stem cells (iPSCs) | Human |
We require the cells/organoids to be washed in PBS to avoid interference with our proprietary lysis and reverse transcription.
The recommended range of input material is in the range of 5’000-50’000 cells.

