Plant BRB-seq service
By leveraging BRB-seq, we not only provide to industrial and academic clients high-quality RNA-seq data, but we also do so with the highest affordability and shortest turnaround times on the market.
The MERCURIUS™ Plant BRB-seq service offers a convenient and streamlined solution for transcriptomics projects of any size. Clients can send us purified plant RNA samples, which are always quality-checked before launching our Plant BRB-seq pipeline. During the process, we always keep clients informed at defined checkpoint so that we can decide together how to best proceed to the next steps.
Next generation sequencing and data pre-processing (including alignment to the genome of choice) are part of our standard service as well. As a result, we provide our clients raw data, sequencing and alignment reports, and gene count matrices which can be used for downstream gene expression analysis.


Step 1
Step 2
Step 3
Step 4
(Nanodrop and Fragment Analyzer)
![]() 1 week
1 week
Step 5
![]() 2 days
2 days
Step 6
(Qubit, Fragment analyzer, shallow sequencing)
![]() 1 week
1 week
Step 7
![]() 1 week
1 week
Step 8
Step 9
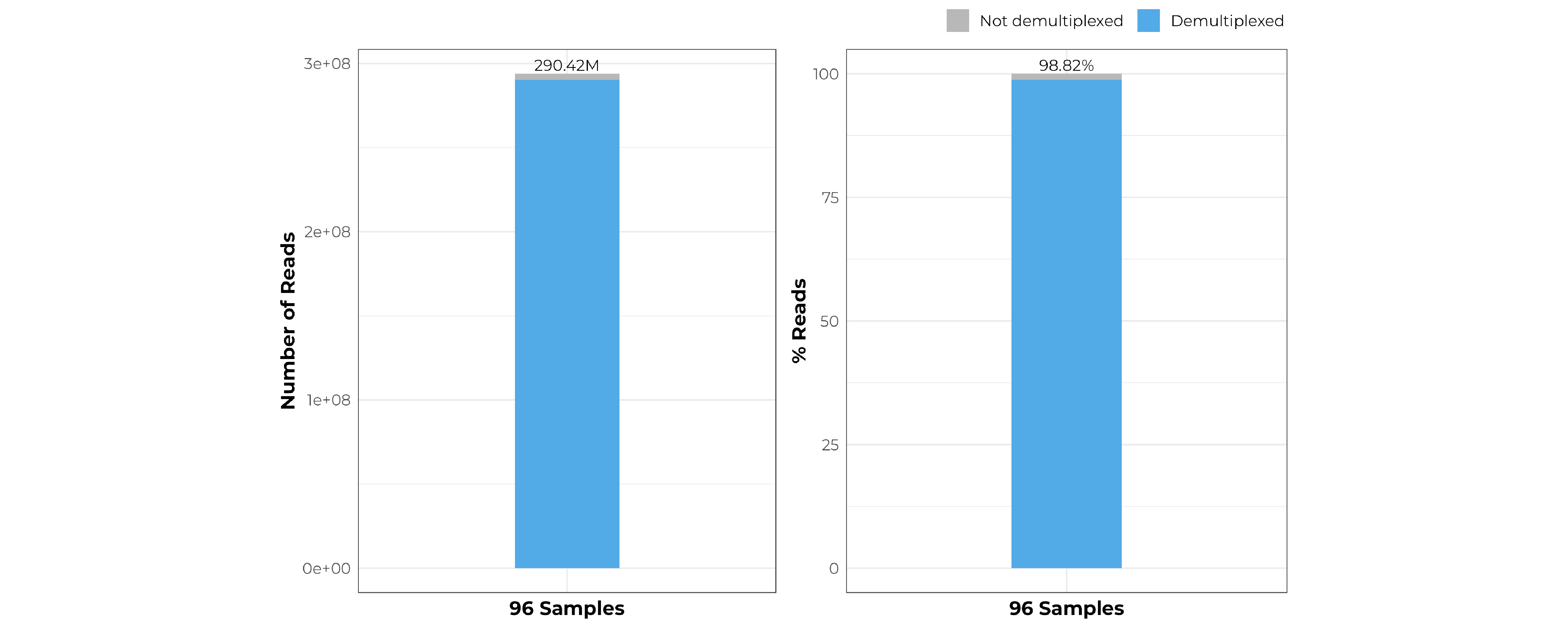
The demultiplexing rate of the pooled 96 samples using MERCURIUS™ Plant BRB-seq library preparation kit is as high as 98%.
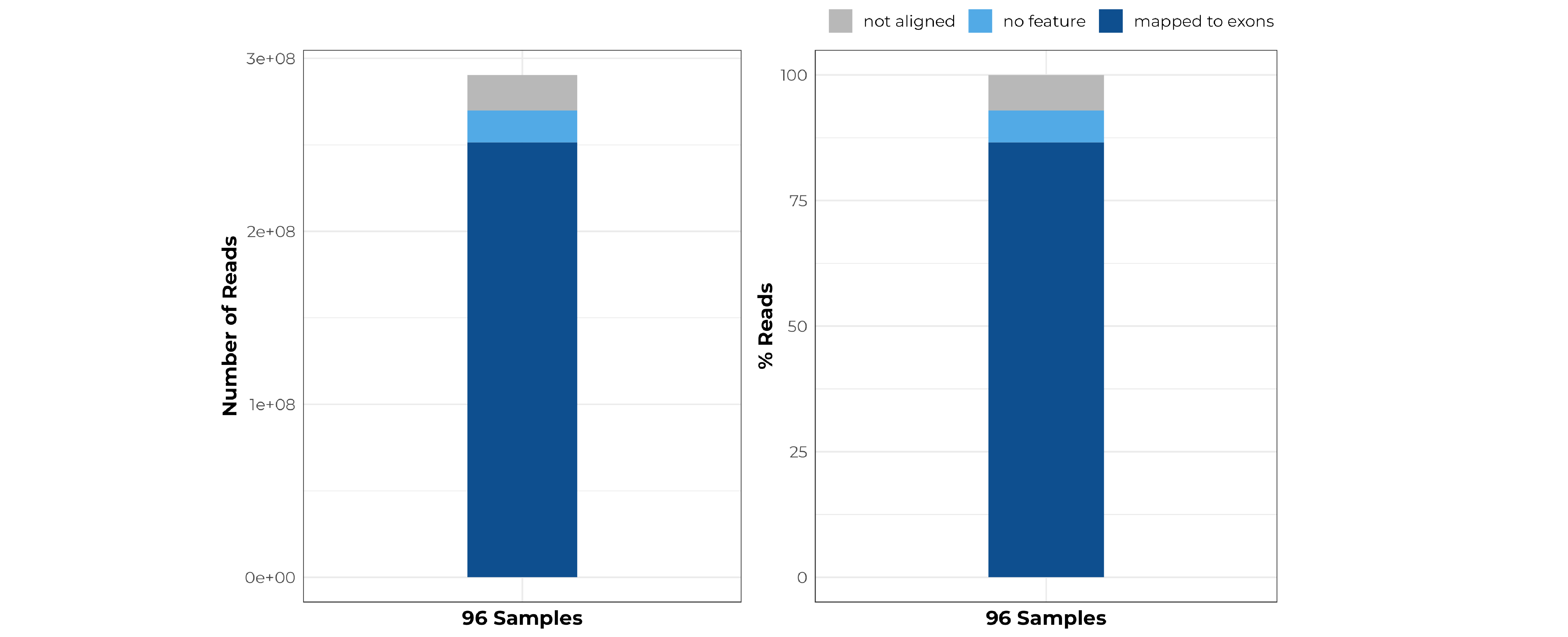
Summary alignment of Arabidopsis data library prepared with the MERCURIUS™ Plant BRB-seq library preparation kit.
.png)
Distribution of the number of detected genes across 96 samples prepared with the MERCURIUS™ Plant BRB-seq library preparation kit. The library was sequenced at an average of 3 million reads.
Number of samples:
96
Reads per sample in demo dataset:
10'000 reads
To have access to the deep-sequenced dataset (3M reads per sample), contact us.
Demo dataset file size:
47.9 MB
Number of samples:
96
Reads per sample in demo dataset:
10'000 reads
To have access to the deep-sequenced dataset (9M reads per sample) contact us.
Demo dataset file size:
83.9 MB
To guarantee high quality data, we normally request that each sample contains at least 12ng/μl of total RNA in at least 15μl.
In addition to total RNA amount, it is important that the samples contain RNA of high integrity (RIN >=6) and are devoid of contaminants (Nanodrop A260/A230 between 1.8 and 2.2).
BRB-seq is 3’-end RNA sequencing method and, as such, requires significantly less sequencing as compared to standard full-length RNA-seq in order to reach accurate gene quantification. We therefore normally recommend to sequence 4 to 5 million reads for each sample, which enables the reliable and unbiased detection of over 18’000 genes.
As part of our standard service pipeline, we align the generated data to the genome of choice, provide a detailed report on the alignment and gene counting statistics and, finally, provide ready-to-use gene count matrices for downstream analysis.
Optionally, we can include differential gene expression analysis.


Explore the latest, relevant publications in the industry to learn more about our technologies.
Book a one-on-one call with one of our RNA experts to discover how we can assist your next project.
Receive email updates on our latest products and services, news and event updates and more. No ads, no spam.