FLASH-seq technology
Ultra-sensitive, full-length and plate-based single-cell and low-input RNA-seq technology for FACS-sorted cells or low-input RNA samples
Ultra-sensitive, full-length and plate-based single-cell and low-input RNA-seq technology for FACS-sorted cells or low-input RNA samples
.png?width=3840&height=2160&name=Firefly%20closeup%20Still%20(1).png)


MERCURIUS™ FLASH-seq provides full-length mRNA transcript coverage even for low-abundance genes, empowering researchers to explore differential gene expression, detect alternative splicing, and analyze isoform diversity—all critical for understanding complex biology, especially in rare cell populations.
The technology is based on the method first published in Nature Biotechnology, which offers unmatched sensitivity and reduces hands-on time compared to Smart-seq2 and Smart-seq3.
The MERCURIUS™ FLASH-seq protocol achieves these improvements in speed and sensitivity by combining the reverse transcription (RT) and cDNA preamplification stages, harnessing a more processive reverse transcriptase with a shortened RT reaction time, and enhancing the template-switching reaction crucial for full-length cDNA synthesis.
Up to 2x more genes detected than other commercially available solutions. Ideal for rare cell capture.
From as low as one cell or 1-100 pg input.
From differential gene expression to splicing variants and isoform detection.
Simply FACS-sort the cells in the wells containing the buffer for complete lysis and efficient reverse transcription.
Enables seamless integration into high-throughput workflows, furter reducing hands-on time and increasing scalability, reproducibility and efficiency.
By mapping gene expression trajectories across individual cells during early development, FLASH-seq allows researchers to study how cells differentiate and form tissues, helping to unravel the complex processes that drive organismal development.
The sensitivity of FLASH-seq makes it efficient even with sub-cellular amounts of RNA (<1 pg) such as exosomes preparations or biopsies of individual cells.
Researchers can use FLASH-seq to evaluate the effect of antisense oligonucleotides (ASOs) on target splicing at single-cell resolution. This enables precise tracking of treatment efficacy across rare or responsive cell subsets within a population.

The number of detected genes in HEK 293T cells processed with different protocols. Reads were downsampled to 500,000 raw reads.
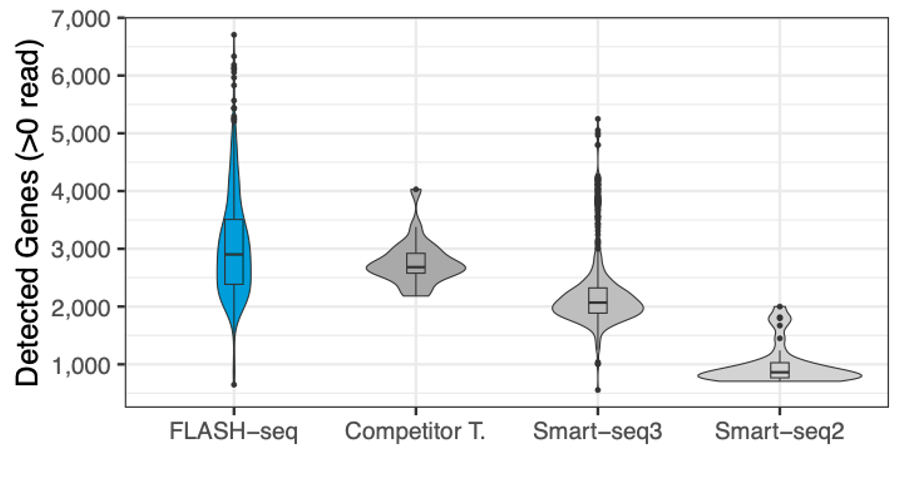
Number of genes detected in human PBMCs, processed with different protocols, and the number of reads downsampled to 125,000 raw reads.
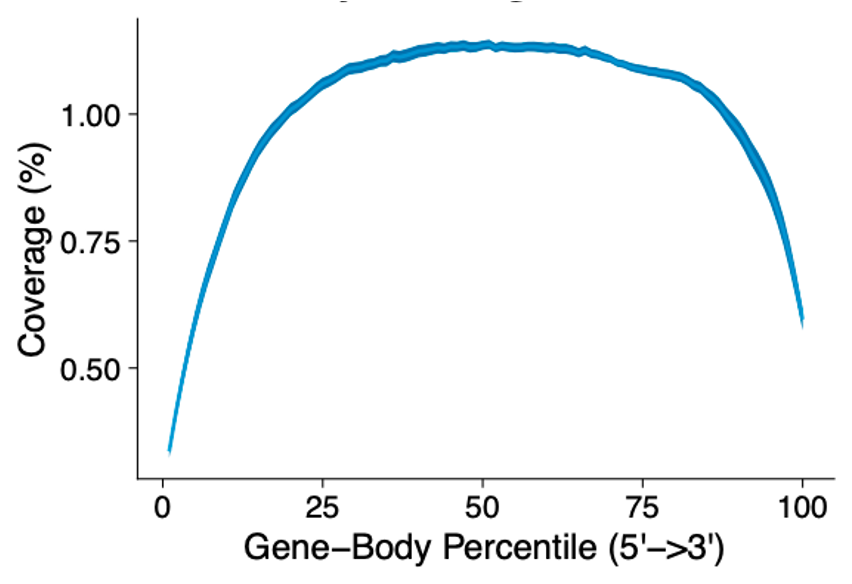
Gene body coverage shows a uniform read distribution across the entire gene body for the FLASH-seq protocol.
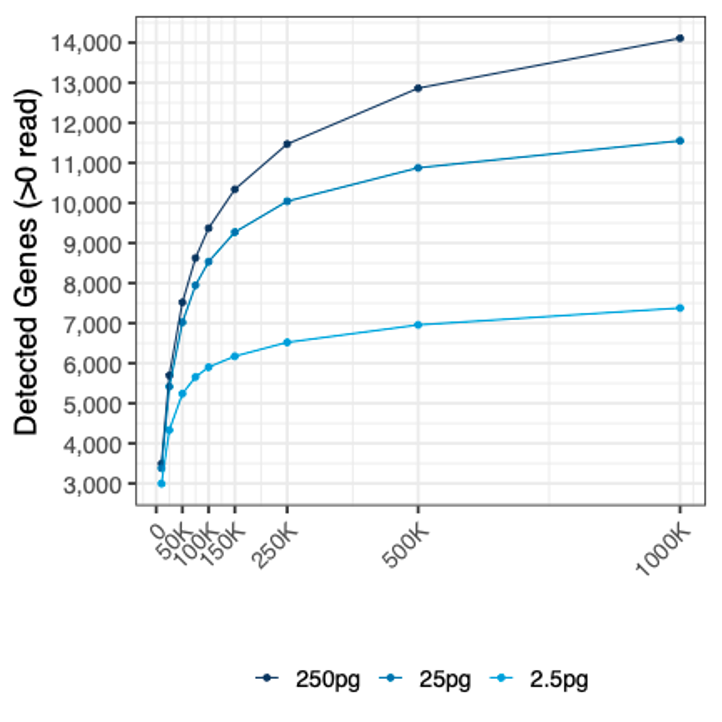
The number of genes detected in HEK 293T cells using different RNA inputs (from 2.5 pg to 250 pg) at different sequencing depths.
All our kits contain all the oligos and enzymes needed to go from FACS-sorted single-cells or low-input RNA samples to sequencing-ready libraries.
MERCURIUS™


MERCURIUS™
MERCURIUS™


MERCURIUS™

With MERCURIUS™ FLASH-seq, we are removing long-standing barriers in single-cell RNA-seq to make full-length, high-resolution transcriptomics truly accessible. Researchers studying rare cells or running high-throughput screens no longer have to compromise between sensitivity, scale, and cost. This launch is a bold step toward accelerating drug discovery and deepening our understanding of disease biology.
Riccardo Dainese, CEO and Co-founder
Check out our blog posts for detailed insights into the FLASH-seq technology or try our Selection Tool to find the best fit for your project.
13 Jan 2026
Maxine Leonardi, Yves Paychère, Felix Naef. Cell-cycle inhibition preserves robust development but rebalances lineages in mouse gastruloids. bioRxiv 2026.01.08.698406
Developmental Biology
13 Jan 2026
Pham, C.N., Castelli, F., Finet, F., Leroy, C., Chollet, C., Chirayath, T.W., Moitra, S., Zarka, M., Ostertag, A., Brial, F., Combes, C., Latourte, A., Bardin, T., Fenaille, F., Richette, P. and Ea, H.K. (2026), Spermidine Reproduces the Anti-Inflammatory Effects of Intermittent Fasting and Prevents Urate and Calcium Pyrophosphate Crystal-Induced Inflammation. Arthritis Rheumatol (2025)
Inflammation
18 Nov 2025
Wu, C., Wang, T., Ghosh, A. et al. MTCH2 modulates CPT1 activity to regulate lipid metabolism of adipocytes. Nat Commun 16, 8831 (2025).
Metabolism
MERCURIUS™ FLASH-seq is a plate-based method that focuses on polyadenylated RNA, which includes most mRNAs. It provides detailed information on gene structure and alternative splicing. Its ability to capture full-length mRNA transcripts and detect low-abundance genes makes it a powerful tool for gene expression profiling.
FLASH-seq protocol was designed to enhance both the sensitivity and efficiency of single-cell mRNA sequencing compared to the most famous plate-based method, Smart-seq2
Read more about the technology in our blog posts: here and here.
We have significantly enhanced the original FLASH-seq method to offer a streamlined workflow and superior data output. This plate-based technology (available in 96- and 384-well formats) features a novel, non-toxic tagmentation buffer and delivers ultra-sensitive gene detection, capturing up to two times more genes compared to other commercially available solutions.
The MERCURIUS™ FLASH-seq protocol is fully compatible with whole FACS-sorted cells.
We do not recommend very large cells, such as the cardiomyocytes, as they are not compatible with the FACS sorting step.
We require the cells to be directly FACS-sorted in the dedicated well plates. The 96- and 384-plates contain the lysis buffer. It is important for the user to sort the cells in the middle of the well. Please refer to the User Guide and the Sample Submission Guidelines for more details.
The average recommended sequencing depth is 250'000 reads/cell.

