Cell lysis module
-
Optimized lysis buffer for complete lysis and efficient reverse transcription.
-
Compatible with DRUG-seq library prep kits.
Massively multiplexed and extraction-free library preparation kits
Compatible with Illumina® and AVITI™ sequencers.
Catalog number | #10841 |
#11041 |
#10851 |
#11051 |
|
Total preps
|
96 | 384 | 384 | 1,536 |
|
Sample multiplexing and plate format
|
96 | 96 | 384 | 384 |
|
Barcoded oligo-dT plates included
|
1 | 4 | 1 | 4 |
|
UDI pairs included
|
4 | 4 | 4 | 4 |
| Documentation | ||||
| Cat ##10841 | ||||
|---|---|---|---|---|
|
Total reactions
|
96 | |||
|
RNA multiplexing format
|
96 | |||
|
UDI pairs included
|
4 | |||
| Cat ##11041 | ||||
|---|---|---|---|---|
|
Total reactions
|
384 | |||
|
RNA multiplexing format
|
96 | |||
|
UDI pairs included
|
4 | |||
| Cat ##10851 | ||||
|---|---|---|---|---|
|
Total reactions
|
384 | |||
|
RNA multiplexing format
|
384 | |||
|
UDI pairs included
|
4 | |||
| Cat ##11051 | ||||
|---|---|---|---|---|
|
Total reactions
|
||||
|
RNA multiplexing format
|
384 | |||
|
UDI pairs included
|
4 | |||
Optimized lysis buffer for complete lysis and efficient reverse transcription.
Compatible with DRUG-seq library prep kits.

Compatible with the MERCURIUS™ product line.
12 UDI pairs.
Flexible dual indexing solutions.
The MERCURIUS™ Extraction-free DRUG-seq kits contain all the oligos and enzymes needed to go from 2D cell cultures to sequencing-ready libraries.
Without pre-amplification, leading to higher mapping and gene detection rates.

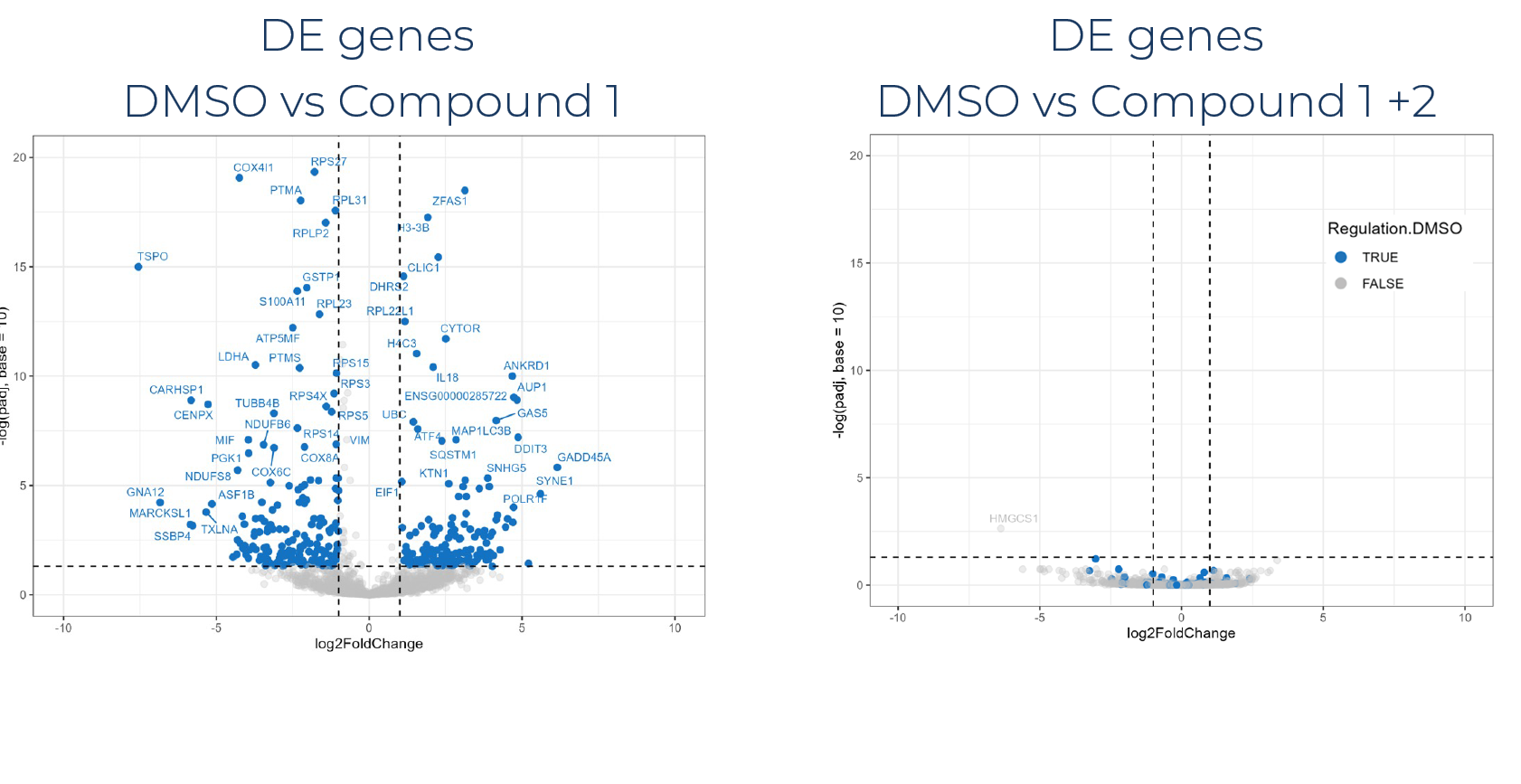
Large-scale analysis of synergistic/antagonistic compound combinations effects
1. Gene detection overview

Gene detection overview of a case study on 12,000 samples and 3,840 compound combinations sequenced at 1M reads per sample.
1. MERCURIUS™ DRUG-seq enables accurate clustering and co-clustering analysis based on unbiased gene expression measurement

2. Detect potential synergistic/antagonistic effects of drug combinations
|
For (application) |
3' mRNA sequencing |
|
For use with (equipment) |
Illumina and AVITI NGS instruments |
|
Species compatibility |
All eukaryotic species |
|
Available formats |
96, 384 preps 1'536 preps |
|
Shipping conditions |
Dry ice |
|
Storage conditions |
-20C |
| Cell line/Tissue | Organism |
| HT1080 SMARCA4 KO | Human |
| hTEC | Human |
| iPSC microglia | Human |
| MCF7 | Human |
| HepG2_IGF1RKO | Human |
| Beas-2B | Human |
| dTHP-1 | Human |
| iPSC derived cardiomyocytes | Human |
| Endothelial | Human |
| Hepatocyte | Human |
| HEPG2 | Human |
| Hepatocyte | Human |
| SF9 | Spodoptera |
| A549 | Human |
| Breast Cancer (MCF7) | Human |
| B lymphoblast MM.1s. | Human |
| Hek293 | Human |
| HeLa Cells | Human |
| U2-OS | Human |
| Hek293 | Human |
| dTHP-1 | Human |
| AsPC-1 | Human |
| PBMC (TCD4) | Human |
| Skeletal muscle (LHCN-M2) | Human |
| HepaRG | Human |
| Macrophage (MV-4-11) | Human |
| PBMCs (Blood) | Human |
| Microglia | Human |
| Fibroblast Like Synoviocyte donor 1502 | Human |
| Fibroblast | Human |
| iPSC-derived neuron | Human |
| Human lung carcinoma epithelial cell line | Human |
| Human breast epithelial adenocarcinoma cell line | Human |
| Human liver hepatocellular carcinoma cell line | Human |
| Human cardiomyocytes derived from induced pluripotent stem cells (iPSCs) | Human |

Large-scale analysis of synergistic/antagonistic compound combinations effects
1. Gene detection overview

Gene detection overview of a case study on 12,000 samples and 3,840 compound combinations sequenced at 1M reads per sample.
1. MERCURIUS™ DRUG-seq enables accurate clustering and co-clustering analysis based on unbiased gene expression measurement

2. Detect potential synergistic/antagonistic effects of drug combinations

|
For (application) |
3' mRNA sequencing |
|
For use with (equipment) |
Illumina and AVITI NGS instruments |
|
Species compatibility |
All eukaryotic species |
|
Available formats |
96, 384 preps 1'536 preps |
|
Shipping conditions |
Dry ice |
|
Storage conditions |
-20C |
| Cell line/Tissue | Organism |
| HT1080 SMARCA4 KO | Human |
| hTEC | Human |
| iPSC microglia | Human |
| MCF7 | Human |
| HepG2_IGF1RKO | Human |
| Beas-2B | Human |
| dTHP-1 | Human |
| iPSC derived cardiomyocytes | Human |
| Endothelial | Human |
| Hepatocyte | Human |
| HEPG2 | Human |
| Hepatocyte | Human |
| SF9 | Spodoptera |
| A549 | Human |
| Breast Cancer (MCF7) | Human |
| B lymphoblast MM.1s. | Human |
| Hek293 | Human |
| HeLa Cells | Human |
| U2-OS | Human |
| Hek293 | Human |
| dTHP-1 | Human |
| AsPC-1 | Human |
| PBMC (TCD4) | Human |
| Skeletal muscle (LHCN-M2) | Human |
| HepaRG | Human |
| Macrophage (MV-4-11) | Human |
| PBMCs (Blood) | Human |
| Microglia | Human |
| Fibroblast Like Synoviocyte donor 1502 | Human |
| Fibroblast | Human |
| iPSC-derived neuron | Human |
| Human lung carcinoma epithelial cell line | Human |
| Human breast epithelial adenocarcinoma cell line | Human |
| Human liver hepatocellular carcinoma cell line | Human |
| Human cardiomyocytes derived from induced pluripotent stem cells (iPSCs) | Human |
Number of samples:
360
Reads per sample in demo dataset:
10'000 reads
To have access to the deep-sequenced dataset (31 GB reads per sample) contact us.
Demo dataset file size:
217 MB
Number of samples:
24
Reads per sample in demo dataset:
10'000 reads
To have access to the deep-sequenced dataset (7.3 M reads per sample) contact us.
Demo dataset file size:
13.3 MB
Number of samples:
24
Reads per sample in demo dataset:
10'000 reads
To have access to the deep-sequenced dataset (7.1 M reads per sample) contact us.
Demo dataset file size:
13.6 MB
Each DRUG-seq or BRB-seq kit contains reagents (including four pairs of Unique Dual Indexing adapters) sufficient for the complete library preparation process for four different BRB-seq pools.
To note, the total number of RNA samples that can be processed with one kit does not exceed the kit specifications; for instance, a 96-samples kit can be used to prepare up-to 96 samples distributed across up-to four different libraries.
The recommended range of input material is in the range of 5’000-50’000 cells.
The only difference between DRUG-seq and standard RNA-seq data analysis is the demultiplexing step, which is used to assign sequencing reads to their sample of origin based on the DRUG-seq barcode sequence.
For a thorough description of DRUG-seq data processing, please refer to the DRUG-seq kit user guide.
The barcode set for your kit is conveniently located on the kit label. Please refer to the label for accurate identification.
For optimal compatibility, ensure that you use the appropriate plate format (e.g., for kits designed for 96 reactions, the 96 well-plate format should be used). This ensures accurate and efficient processing of your samples. If you have any further questions or concerns, please contact our support team for assistance by email or using our live chat tool.



Explore the latest, relevant publications in the industry to learn more about our technologies.
Let us do it for you. Our team delivers raw sequencing data (fastq files), gene count matrices and analysis report files. A cost-efficient option suitable for projects of all sizes.
Book a one-on-one call with one of our RNA experts to discover how we can assist your next project.
Receive email updates on our latest products and services, news and event updates and more. No ads, no spam.