MERCURIUS™
FFPE-seq service
Massively multiplexed full-length total RNA-seq for heavily degraded samples.
MERCURIUS™
Massively multiplexed full-length total RNA-seq for heavily degraded samples.


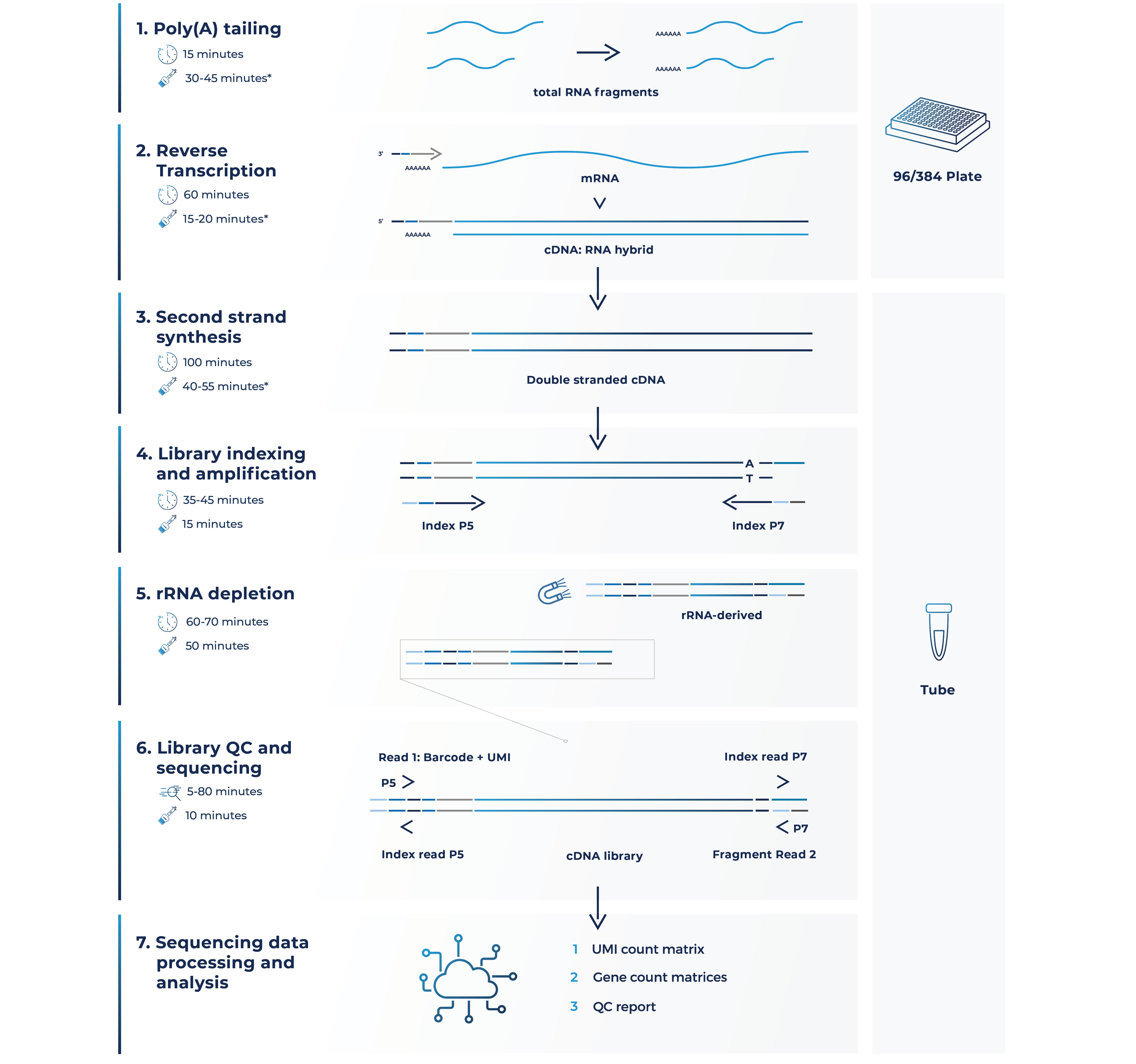


Step 1
Step 2
Step 3
Step 4
![]() 2 days
2 days
Step 5
(Qubit, Fragment analyzer, shallow sequencing)
![]() 1 week
1 week
Step 6
![]() 1 week
1 week
Step 7
Step 8
Read more about the FFPE-seq on our blog.

Genome browser view of the RNA-seq reads mapped to several human genes, using different library preparation protocols. The FFPE-seq shows comparable coverage as the NEB Ultra II, while the BRB-seq protocol is limited only to the 3' end genomic region.
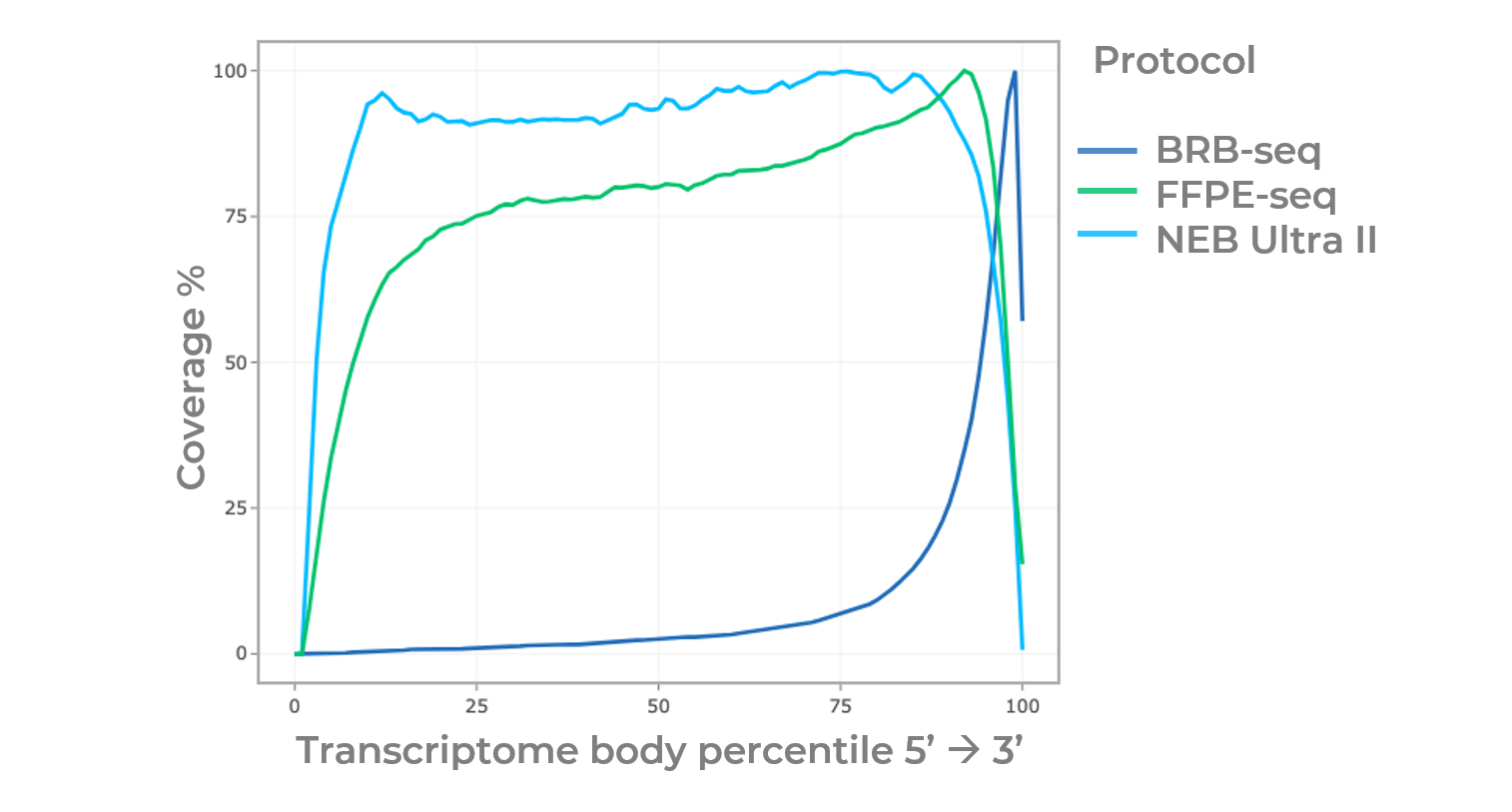
Gene body coverage from FFPE-seq, BRB-seq, and NEB Ultra II. The FFPE-seq shows a similar coverage profile as the NEB Ultra II. By contrast, BRB-seq reads preferentially mapped to the 3'end.
We recommend that each sample contains 500ng of total RNA to guarantee high-quality data.
The service is validated with the purified RNA samples from various source and RIN values ranging from 1 to 10.
We normally recommend sequencing between 10-20 million reads for each sample.
As part of our standard service pipeline, we align the generated data to the genome of choice, provide a detailed report on the alignment and gene counting statistics and, finally, provide ready-to-use gene count matrices for downstream analysis.
Optionally, we can include differential gene expression analysis.


Explore the latest, relevant publications in the industry to learn more about our technologies.
Book a one-on-one call with one of our RNA experts to discover how we can assist your next project.
Receive email updates on our latest products and services, news and event updates and more. No ads, no spam.