DRUG-seq service
DRUG-seq boosts screening efforts based on gene expression profiling by offering a unique RNA-seq service that is highly sensitive, extraction-free, massively multiplexed and extremely cost-effective.
DRUG-seq boosts screening efforts based on gene expression profiling by offering a unique RNA-seq service that is highly sensitive, extraction-free, massively multiplexed and extremely cost-effective.




Step 1
Step 2
Step 3
![]() 2 days
2 days
Step 4
(Qubit, Fragment analyzer, shallow sequencing)
![]() 1 week
1 week
Step 5
![]() 1 week
1 week
Step 6
Step 7
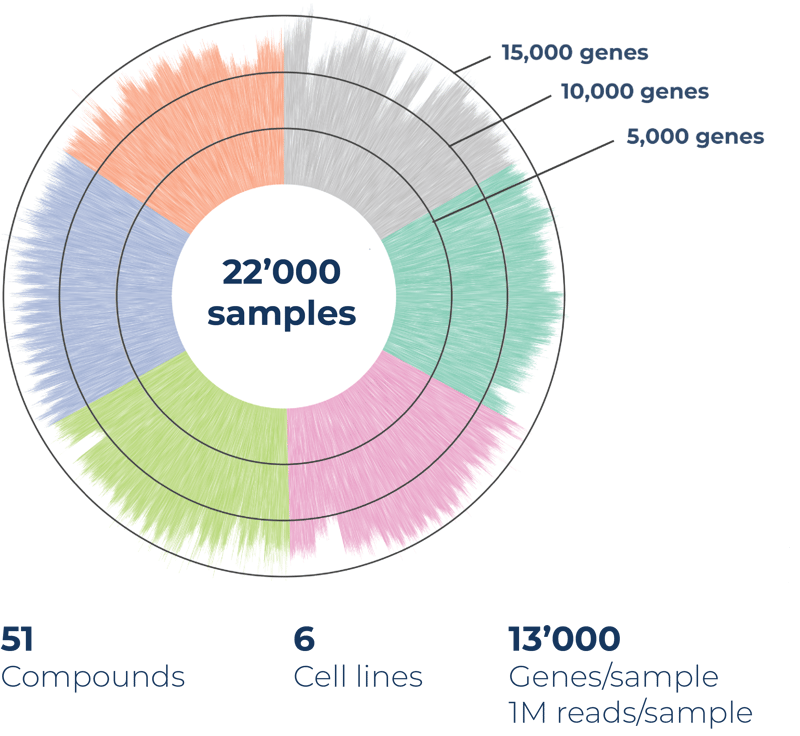
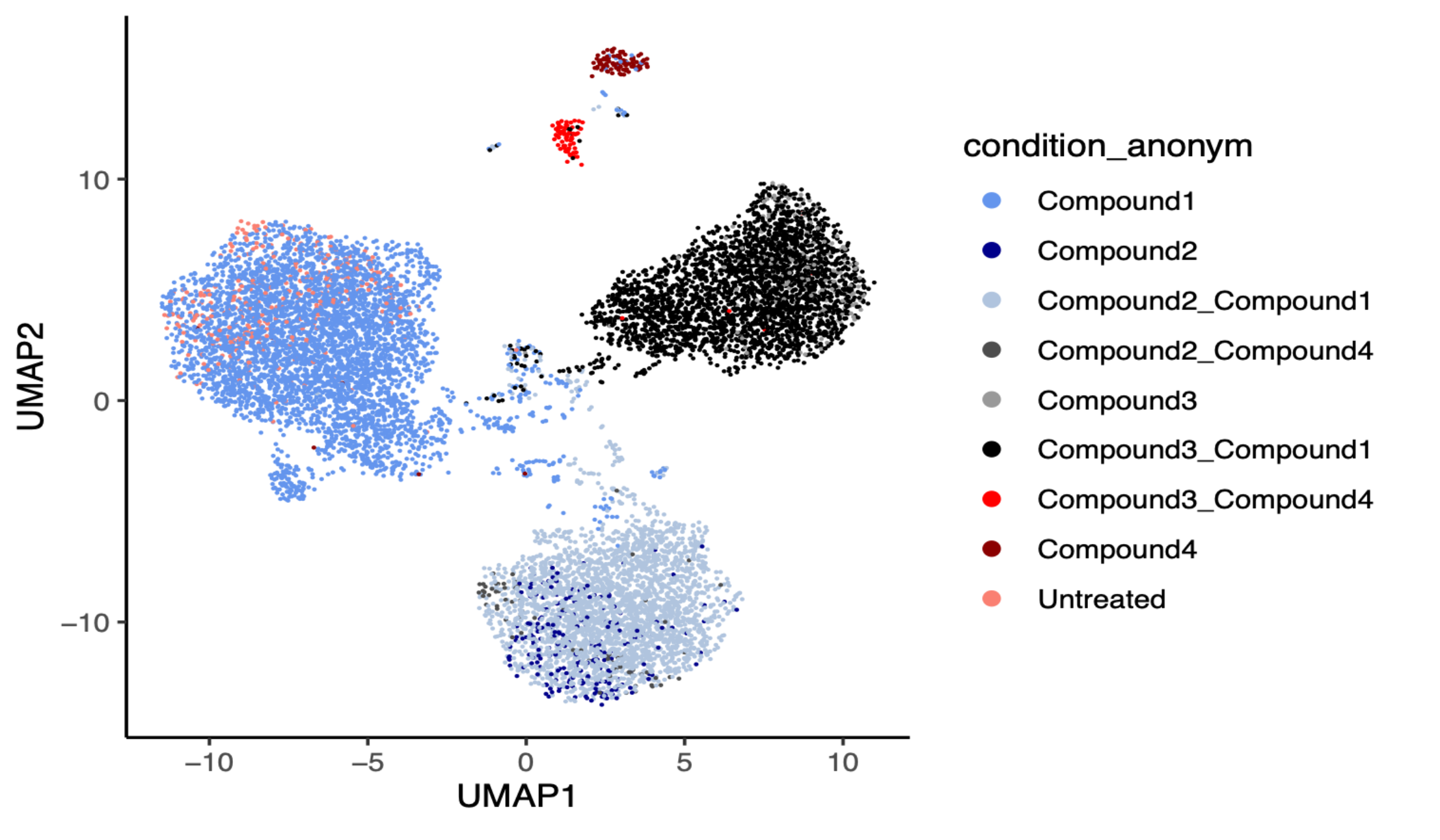
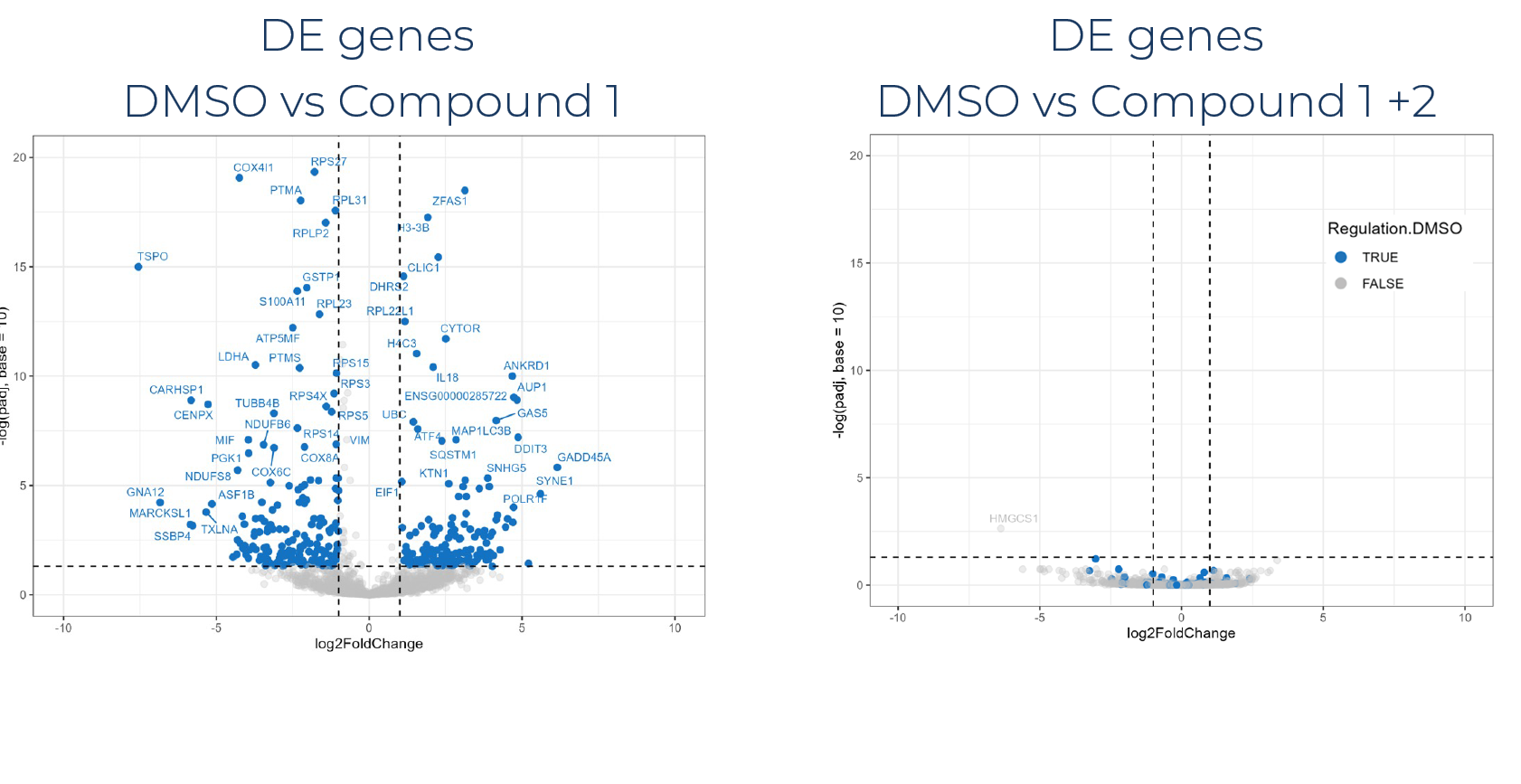
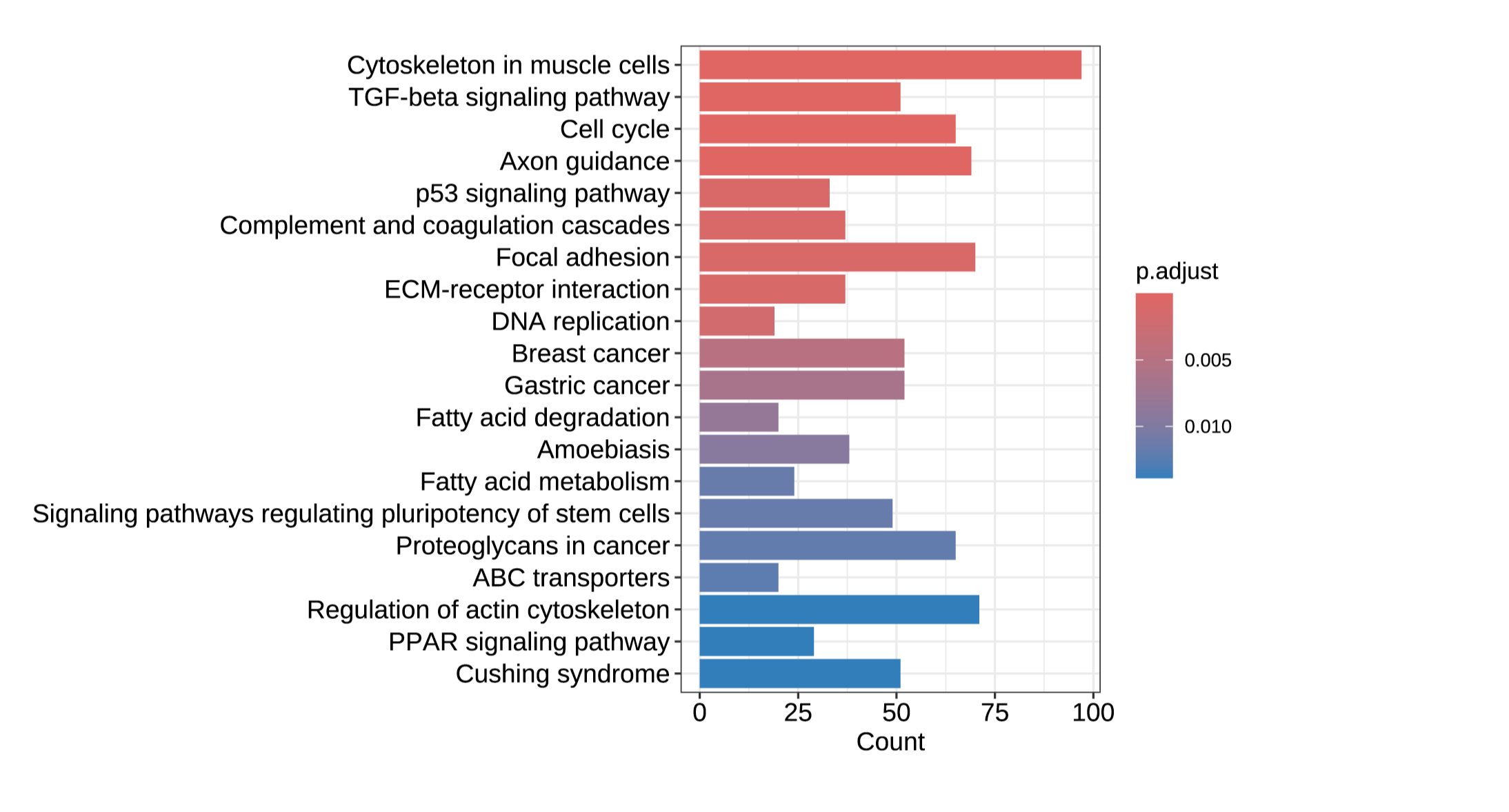

Number of samples:
24
Reads per sample in demo dataset:
10'000 reads
To have access to the deep-sequenced dataset (7.3 M reads per sample) contact us.
Demo dataset file size:
13.3 MB

Number of samples:
65
Reads per sample in demo dataset:
10'000 reads
To have access to the deep-sequenced dataset (3.9 M reads per sample) contact us.
Demo dataset file size:
4.76 MB

Number of samples:
24
Reads per sample in demo dataset:
10'000 reads
To have access to the deep-sequenced dataset (7.1 M reads per sample) contact us.
Demo dataset file size:
13.6 MB
To guarantee high quality data, we normally request that each sample contains 15k-50k cells/well for 96 well-plate or 2k-10k cells/well for 384 well-plate. The minimum number of cells per pool should be 80k.
MERCURIUS™ DRUG-seq can be performed on various sample types, including: Cell lines, Primary cells, Organoids or Spheroids.
Validated cells lines
| Cell line/Tissue | Organism |
| HT1080 SMARCA4 KO | Human |
| hTEC | Human |
| iPSC microglia | Human |
| MCF7 | Human |
| HepG2_IGF1RKO | Human |
| Beas-2B | Human |
| dTHP-1 | Human |
| iPSC derived cardiomyocytes | Human |
| Endothelial | Human |
| Hepatocyte | Human |
| HEPG2 | Human |
| Hepatocyte | Human |
| SF9 | Spodoptera |
| A549 | Human |
| Breast Cancer (MCF7) | Human |
| B lymphoblast MM.1s. | Human |
| Hek293 | Human |
| HeLa Cells | Human |
| U2-OS | Human |
| Hek293 | Human |
| dTHP-1 | Human |
| AsPC-1 | Human |
| PBMC (TCD4) | Human |
| Skeletal muscle (LHCN-M2) | Human |
| HepaRG | Human |
| Macrophage (MV-4-11) | Human |
| PBMCs (Blood) | Human |
| Microglia | Human |
| Fibroblast Like Synoviocyte donor 1502 | Human |
| Fibroblast | Human |
| iPSC-derived neuron | Human |
| Human lung carcinoma epithelial cell line | Human |
| Human breast epithelial adenocarcinoma cell line | Human |
| Human liver hepatocellular carcinoma cell line | Human |
| Human cardiomyocytes derived from induced pluripotent stem cells (iPSCs) | Human |
DRUG-seq is 3’-end RNA sequencing method and, as such, requires significantly less sequencing as compared to standard full-length RNA-seq in order to reach accurate gene quantification. We therefore normally recommend to sequence 1 to 10 million reads for each sample, which enables the reliable and unbiased detection of over 18’000 genes.
As part of our standard service pipeline, we align the generated data to the genome of choice, provide a detailed report on the alignment and gene counting statistics and, finally, provide ready-to-use gene count matrices for downstream analysis.


Explore the latest, relevant publications in the industry to learn more about DRUG-seq.
Experience reliable, cost-effective, and scalable products that deliver high-quality data for large projects.
Book a one-on-one call with one of our RNA experts to discover how we can assist your next project.