Accurately measuring gene expression is fundamental to modern drug discovery and development, enabling researchers to evaluate cellular responses to potential therapeutic compounds. One of the widely used technologies for this purpose is the TaqMan™ gene expression assay, a real-time PCR-based method that offers high sensitivity, specificity, and reproducibility.
In this article, we delve into the science behind the TaqMan assay, break down its mechanism, and explain how fluorescent probe cleavage enables gene expression measurement. We will also explore the various analysis and assay options available, from predesigned to custom and high-throughput formats, catering to different research needs.
The Science Behind TaqMan Gene Expression Assays
TaqMan gene expression assays are a powerful tool for quantifying RNA with high specificity and sensitivity using real-time PCR technology [1]. Each assay consists of a single tube containing two PCR primers and a 5’ fluorescent FAM™ dye-labelled probe that includes a 3’ nonfluorescent quencher and minor groove binder. The primers and probe bind specifically to their complementary target sequences, enabling the precise measurement of gene expression levels (Fig. 1).
The assay relies on the 5' nuclease activity of Taq polymerase to cleave the probe during PCR amplification. When the probe is intact, the fluorophore is in proximity to the nonfluorescent quencher, and fluorescence is suppressed. However, upon binding of the primers and probe at the correct genomic location, the Taq polymerase extends the DNA strand and cleaves the probe, separating the reporter dye from the quencher. This results in an increase in fluorescence that is directly proportional to the amount of target RNA present, enabling accurate quantification.
For optimal results, researchers often use purified total RNA extracted from cells or tissues, ensuring it is free of contaminants such as genomic DNA, proteins, or enzymatic inhibitors. Researchers can also use the assays directly with cell lysates, eliminating the need for RNA purification. The TaqMan Cells-to-CT™ Express Kit is specifically designed for this purpose, allowing researchers to perform real-time RT-qPCR directly from cultured cells [2].
Step-by-Step Mechanism
1. Probe Design & Components:
- The TaqMan probe is a short, single-stranded oligonucleotide complementary to the target sequence.
- It carries a fluorescent reporter dye at the 5' end and a quencher at the 3' end.
- When intact, the quencher suppresses fluorescence from the reporter dye.
2. Annealing & Extension:
- During the annealing phase, the probe binds to its complementary target sequence.
- The forward and reverse primers also anneal to the template DNA.
- As Taq polymerase extends the primers, it encounters the probe.
3. Probe Cleavage & Fluorescence:
- The 5' nuclease activity of Taq polymerase cleaves the probe.
- This separates the reporter dye from the quencher, allowing fluorescence emission to occur.
- The fluorescence intensity increases proportionally to the amount of amplified DNA.
4. Real-Time Quantification:
- A fluorescence detector measures the emitted signal at each PCR cycle.
- The signal correlates with the amount of target nucleic acid present
- This enables precise quantification of gene expression levels.
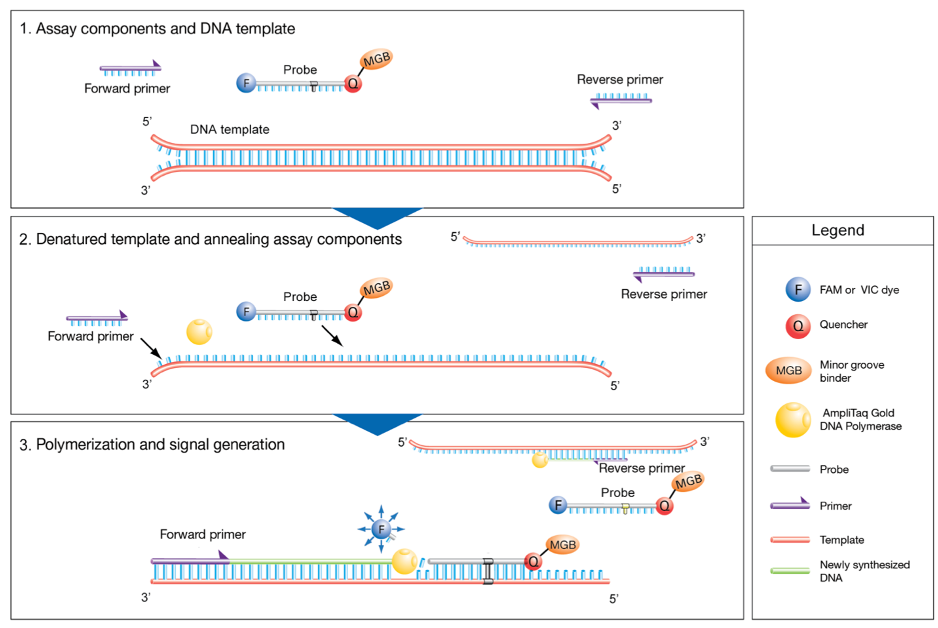
Figure 1. Sample qPCR workflow at a glance using TaqMan assays (taken from [1])
Behind the Workflow: Generating, Collecting, and Interpreting Data in TaqMan Assays
Accurate data collection and thoughtful interpretation are critical to the success of TaqMan assays. Real-time PCR instruments track the amplification of genetic material cycle by cycle, capturing fluorescence signals that reflect DNA accumulation in real time. A key metric derived from this process is the cycle threshold (Ct), which represents the point at which the fluorescent signal exceeds the background noise.
The Ct value serves as an indirect measure of the starting amount of target nucleic acid: the lower the Ct, the higher the initial concentration. Interpreting these values requires care, as they can be influenced by both biological variability and technical factors. Including appropriate positive and negative controls helps ensure that observed changes in expression are meaningful and not artifacts.
Positive Control Examples:
GAPDH or ACTB (beta-actin)
- These are housekeeping genes that are stably and consistently expressed in most cell types.
- They are often included to confirm that
- cDNA synthesis was successful.
- The qPCR reaction is working as expected.
- There are no inhibitors in the reaction mix.
Negative Control Examples:
1. No Template Control (NTC)
- This refers to a reaction that contains all reagents except the cDNA template.
- It should be included to detect contamination (e.g., in reagents, pipetting). There should be no amplification in this control.
2. No Reverse Transcriptase Control (No-RT)
- This control processes RNA like other samples but without reverse transcriptase.
- It ensures the signal isn’t coming from genomic DNA. No amplification should be seen if primers are exon-exon spanning.
Key Equipment and Analytical Tools
Real-time PCR instruments known as thermocyclers are essential for performing TaqMan assays. These systems detect DNA amplification in real time using fluorescence, offering highly sensitive and quantitative results. Key features include precise temperature control and the ability to run multiple samples in parallel.
Data Analysis Approaches
Specialized software can help analyze fluorescence data generated by real-time PCR machines. These tools calculate gene expression levels, often relative to stable housekeeping genes, and may include built-in statistical analysis and data visualization features, helping researchers extract meaningful insights from their experiments. However, some platforms can be complex and may require user training to fully leverage their capabilities.
Here are some tried and tested ways to analyze your TaqMan gene expression assay data using Ct values [4].
- Absolute quantification - determines gene expression usually by relating the Ct signal to a standard curve of known template concentrations.
- Relative quantification - relates the Ct signal of the target transcript in a treatment group to that of another sample, such as an untreated control. The 2-ΔΔCt method is a popular method for analyzing the relative changes in gene expression from real-time quantitative PCR experiments.
Different Assay Options
Users can select from predesigned or custom TaqMan assays, which are available in several formats to accommodate diverse research [3].
- Predesigned Assays: Covering over 2.8 million targets across more than 30 species, these assays provide broad applicability without requiring optimization. The assays come pre-formulated in a single tube containing a labeled TaqMan probe, along with forward and reverse primers.
- Custom Assays: Researchers can design assays tailored to specific target sequences of any coding or non-coding organism, offering flexibility for unique experimental requirements. This process typically takes from 2 to 5 weeks, and the minimum order is 10 plates. This option is available in various formats, including 96-well plates, 384-well plates, and OpenArray plates, catering to both standard and high-throughput applications [2].
Carefully designing your specific target sequences is key to a successful TaqMan assay. One key issue to avoid is primer-dimer formation, which often occurs due to suboptimal primer sequences. Ensuring that primers and probes lack significant homology is also essential to prevent non-specific amplification.
Additionally, reagent concentrations must be properly optimized—too much or too little can compromise sensitivity and accuracy. Attention to these details helps ensure reliable and reproducible gene expression results.
Applications of TaqMan Gene Expression Assays
TaqMan assays are widely used across molecular biology and medical research, valued for their precision and reliability in measuring gene expression. Two key areas of application are gene expression profiling and clinical diagnostics.
Gene Expression Profiling
TaqMan assays enable researchers to study gene activity across different conditions, shedding light on regulatory mechanisms in development, disease, and cellular function. Their high sensitivity and specificity make them ideal for detecting even low-abundance transcripts, providing insight into subtle biological changes.
With multiplexing capabilities and a broad dynamic range, TaqMan assays can simultaneously quantify multiple genes with accuracy, crucial for small expression studies and high-throughput workflows.
Clinical Diagnostics
In clinical settings, TaqMan assays play a key role in identifying disease markers and informing treatment strategies. Their precision and reproducibility make them especially useful for:
- Cancer Biomarkers: Assays can detect tumor-associated genes with high specificity, aiding in diagnosis, prognosis, and monitoring treatment response. Despite their utility, the development of customized assays can be costly, which may impact accessibility.
- Infectious Disease Detection: The assays allow for rapid and accurate identification of pathogens, distinguishing between closely related strains—a valuable feature in managing infectious outbreaks. Real-time results enable faster decision-making, although assay sensitivity can be affected by poor sample quality or low pathogen load.
Final Thoughts: When to Consider Alternative Approaches
Research using TaqMan assays involves the careful selection of sample types, control of variables that could influence gene expression, and adherence to standardized workflows. Real-time PCR analysis tools are also employed to enable precise quantification and robust analysis of expression data.
While TaqMan gene expression assays have served as a gold standard for targeted transcript analysis for decades, their limited scope and predefined panels can create bottlenecks in high-throughput or exploratory research. In studies where the full transcriptional response to a compound or genetic perturbation is unknown, or where flexibility and scale are essential, more comprehensive solutions are needed.
Unbiased, whole-transcriptome screening approaches, like MERCURIUS™ DRUG-seq, offer a powerful alternative. By enabling scalable, cost-efficient, and data-rich profiling across the entire transcriptome, directly from cell lysates, MERCURIUS™ DRUG-seq opens new possibilities for discovery-driven research and high-throughput screening campaigns alike.
References
- Thermo Fisher Scientific. TaqMan™ Gene Expression Assays Brochure (PDF)
- https://www.thermofisher.com/order/catalog/product/A57987
- Thermo Fisher Scientific.Custom TaqMan™ Gene Expression Assays. Accessed online
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. methods. 2001 Dec 1;25(4):402-8.