Whole blood RNA samples contain high levels of globin RNAs which reduce the sensitivity to detect genes from the rest of the blood transcriptome (Debey et al., 2006; Shin et al., 2014). Numerous globin depletion methods have been developed to block these transcripts before RNA-seq library preparation and sequencing (Jang et al., 2020). This ensures maximum sensitivity to detect relevant and interesting gene candidates.
In this article we put MERCURIUS™ Blood BRB-seq (Alithea Genomics) and GLOBINclear™-Human kit (ThermoFisher) head-to-head to see how these globin depletion methods compare.
Blood BRB-seq performs as well as GLOBINclear™
To determine if MERCURIUS™ Blood BRB-seq blocks globin genes to a level comparable to GLOBINclear™ we performed pre-treatment of isolated whole blood RNA with each globin depletion product, followed by BRB-seq.
When RNA was not pre-treated with globin blockers, only 12% of reads mapped to non-globin genes (Fig. 1A). In contrast, over 75% of all reads mapped to non-globin genes when RNA was treated with either MERCURIUS™ Blood BRB-seq or GLOBINclear™ (Fig. 1A).
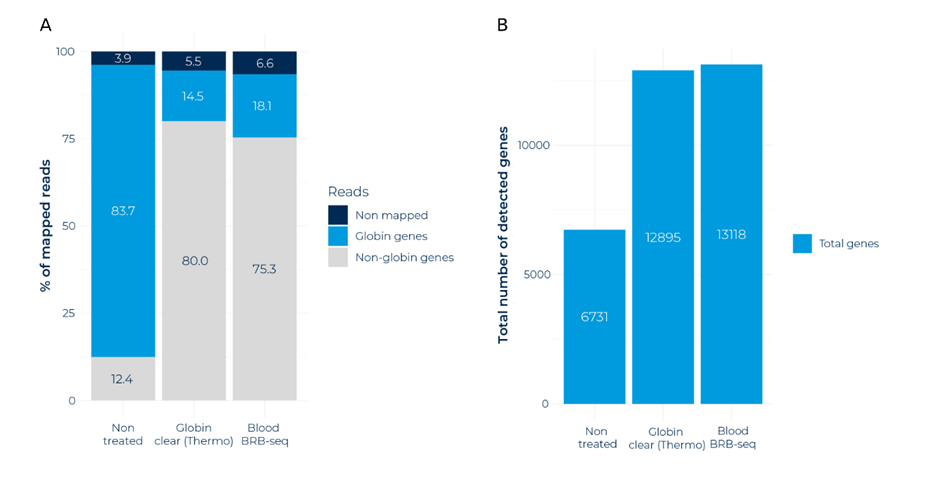
Figure 1. GLOBINclear™ and MERCURIUS™ Blood BRB-seq are highly effective at reducing globin transcripts (A) Stacked bar chart indicating the percentage of mapped reads from blood RNA samples without globin depletion and with globin depletion performed using GLOBINclear™-Human Kit and the MERCURIUS™ Blood BRB-seq method. (B) Number of genes detected across the three different conditions.
Therefore, the integrated human globin blocker from Alithea Genomics used in MERCURIUS™ Blood BRB-seq is as effective as GLOBINclear™. It reliably reduces the amount of globin transcripts in sequencing experiments.
Importantly, the removal of globin RNAs increased the amount of expressed genes detected (Fig. 1B). Only 7000 expressed genes were detected in non-treated samples but this increased to around 13000 genes after treatment with either globin blocker reagent (Fig. 1B).
Therefore, pre-treatment of whole blood RNA with MERCURIUS™ Blood BRB-seq or GLOBINclear™ dramatically increases the number of genes detected by BRB-seq and is a vital preparatory step to ensure rich, globin-free data.
Both treatments specifically target globin genes
Next, we used the MERCURIUS™ Blood BRB-seq and the GLOBINclear™ BRB-seq globin depleted libraries to see if only globin genes were blocked or if other genes were also affected.
For both techniques, globin depletion was effective and specific to the target globin genes (Fig. 2).
Globin genes, such as hemoglobin subunit beta and alpha (HBB and HBA), remained highly expressed in non-treated libraries. Importantly, their expression was dramatically reduced in both treated libraries, thanks to effective blocking of globin transcripts (Fig. 2).
Other genes were not affected by globin blocking treatment (Fig. 2). Non-globin gene expression was highly correlated for non-treated and treated libraries with both techniques (Fig. 2).
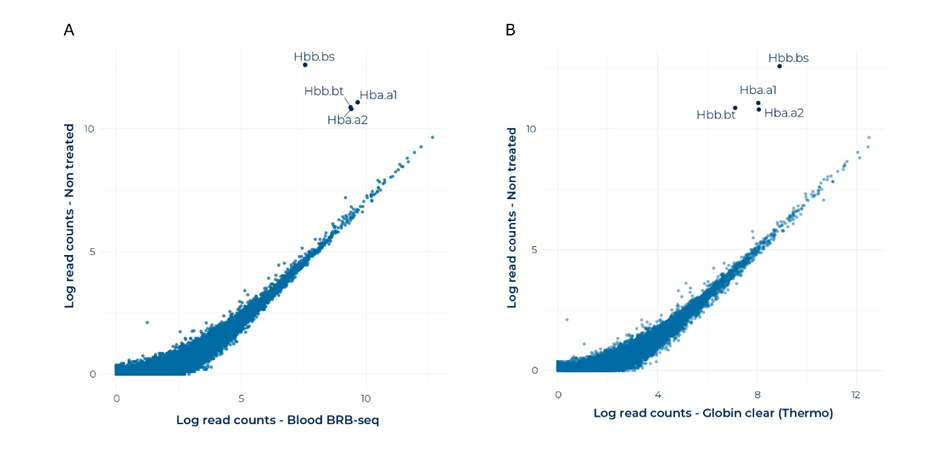
Figure 2. Globin depletion is effective and specific to the target globin genes, with negligible off-target effects. Correlation plots between mapped reads in non-treated libraries and (A) MERCURIUS™ Blood BRB-seq and (B) GLOBINClear™-treated libraries.
MERCURIUS™ Blood BRB-seq and GLOBINclear™ are highly effective
These experiments show that the globin depletion method used in MERCURIUS™ Blood BRB-seq is as highly effective as the gold standard GLOBINclear™ technology.
One advantage of MERCURIUS™ Blood BRB-seq is that treatment with human globin blocker is seamlessly integrated into the pipeline as standard and requires no additional reagents.
Alithea Genomics offer a fully integrated blood sequencing pipeline, including RNA extraction from common blood RNA storage tubes, library preparation, sequencing and final data analysis.
To find out more about how MERCURIUS™ Blood BRB-seq can help in your study, please contact us at info@alitheagenomics.com.
References
- Debey, S., Zander, T., Brors, B., Popov, A., Eils, R. and Schultze, J.L., 2006. A highly standardized, robust, and cost-effective method for genome-wide transcriptome analysis of peripheral blood applicable to large-scale clinical trials. Genomics, 87(5), pp.653-664.
- Jang, J.S., Berg, B., Holicky, E., Eckloff, B., Mutawe, M., Carrasquillo, M.M., Ertekin-Taner, N. and Cuninngham, J.M., 2020. Comparative evaluation for the globin gene depletion methods for mRNA sequencing using the whole blood-derived total RNAs. BMC genomics, 21(1), pp.1-9.
- Shin, H., Shannon, C.P., Fishbane, N., Ruan, J., Zhou, M., Balshaw, R., Wilson-McManus, J.E., Ng, R.T., McManus, B.M., Tebbutt, S.J. and PROOF Centre of Excellence Team, 2014. Variation in RNA-Seq transcriptome profiles of peripheral whole blood from healthy individuals with and without globin depletion. PLoS One, 9(3), p.e91041.


