MERCURIUS™
DRUG-seq
- RNA extraction-free
- 3' mRNA protocol
- Unbiased transcriptome profiling
- Ideal for large-scale compound screening
- Turnaround time: from 2 weeks

We offer a variety of scalable RNA-seq library preparation, sequencing and analysis services that combine full-transcriptome coverage with unmatched cost-efficiency and the shortest turnaround times on the market.
Whether you are looking to generate and analyze data from tens to thousands of samples, our next-generation RNA-seq technologies are designed to fit your research needs and your timeline.
At Alithea Genomics, we specialize in both bulk and single-cell transcriptomics, offering scalable, high-quality RNA sequencing solutions for a wide range of research needs.
Ready to get started? Tell us about your project, and we will provide a personalized quote based on your specific needs and research goals.
Every service project begins with a complimentary consultation, because each study is unique.
Our expert team of scientists will work closely with you to understand your research goals, sample requirements, experimental design, and what data analysis is needed.
This collaborative approach ensures the most suitable methods are chosen from the start, helping you move forward with confidence and clarity.
At Alithea Genomics, we combine advanced automation with cutting-edge technologies to process your samples efficiently.
Our expertise in high-throughput transcriptomics allows us to handle diverse sample types, including cell lysates and low-quality inputs, without compromising on data quality.
Leveraging years of experience in bulk RNA sequencing, our scientists oversee every step of the process within our specialized facility.
With robust quality control embedded throughout the workflow, we ensure that your samples are treated with the care and precision required for downstream success.
Every project includes a preliminary data analysis report designed to accelerate your research.
Our expert bioinformatics team uses proprietary pipelines to provide a clear, concise summary of key results and quality metrics.
For deeper insights, we offer tailored analytical support, ranging from clustering and gene set enrichment to data integration and the generation of publication-ready data.
Whether you are preparing for your next experiment or your next paper, our team is here to help translate complex data into meaningful discoveries.
Unlock deeper insights from your RNA-seq data with our tailored data analysis services.
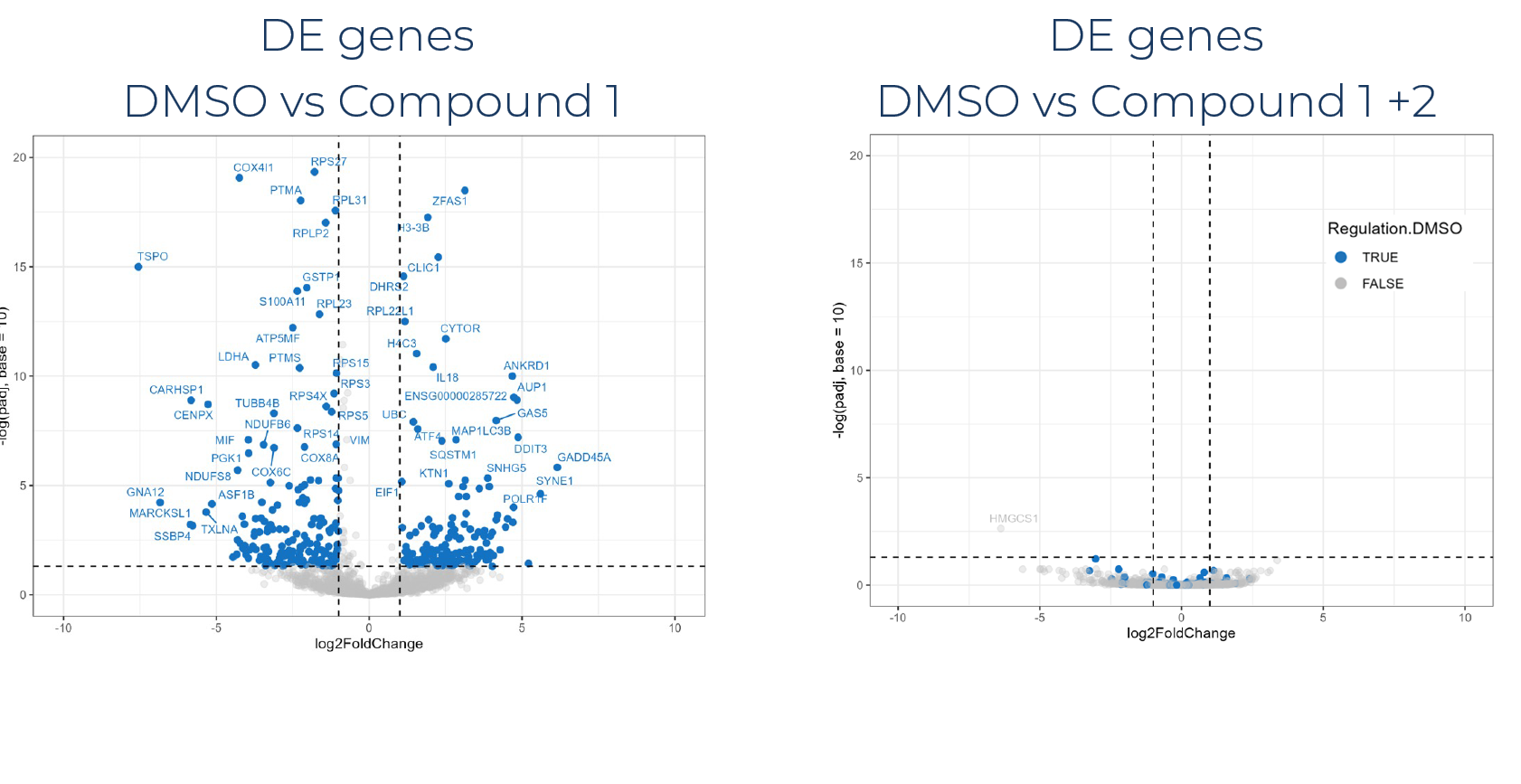
Our preliminary data analysis can be accompanied by publication-ready visualizations such as volcano plots, clustered heatmaps, and our signature barplot wheels.



The DRUG-seq platform enabled us to systematically profile compound-induced transcriptional responses at scale, generating a rich dataset that helped prioritize testable hypotheses. The team at Alithea provided outstanding technical guidance throughout, including support in developing a customized analysis pipeline tailored to our needs. The turnaround time was fast, the process seamless, and the collaboration highly productive. We would be glad to work together again.
Mikołaj Słabicki, Ph.D. Principal Investigator at the MGH Krantz Family Center for Cancer Research. Assistant Professor of Medicine at Harvard Medical School. Affiliate Faculty Member at the Broad Institute of MIT and Harvard
DRUG-seq has completely transformed how I approach and think about transcriptomic screening. It's fast, scalable, and incredibly cost-effective—perfect for profiling hundreds of compounds in parallel. In my experience, it’s one of the most powerful tools for uncovering mechanisms of action and capturing transcriptional signatures at scale. I've used DRUG-seq to profile different cells treated under time points, dose responses, combination treatments, and a variety of complex cellular models, from cardiomyocytes to organoids—and it consistently delivers high-quality, actionable data. The ability to pool samples early without compromising data quality truly makes it a game-changer.
Andrea Hadjikyriacou, Ph.D. Principal Scientist I, Novartis Biomedical Research
BRB-seq was a great way for us to multiplex large numbers of RNA samples for sequencing. We wanted a gene expression curve with a tight temporal resolution, thus, many time points for several conditions. We are very happy with the quality of the results and with the cost benefit.
Dr. Nuno Miguel Luis CNRS Researcher
BRB-seq has proven to be an excellent method for multiplexing a large array of RNA samples for sequencing. Our team sought to generate gene expression profiles to enable our research on possible neurodegeneration therapeutic targets, requiring multiple samples for various experimental conditions. The outcomes have exceeded our expectations in terms of both result quality and cost-effectiveness, leaving us thoroughly satisfied and impressed.
Dr. Emmanouil Metzakopian VP of Research and Development, bit.bio
Check out our blog posts for detailed insights into our services, or read our overview to get the big picture. Still unsure?
Try our Service Selection Tool to find the best fit for your project.
13 Jan 2026
Maxine Leonardi, Yves Paychère, Felix Naef. Cell-cycle inhibition preserves robust development but rebalances lineages in mouse gastruloids. bioRxiv 2026.01.08.698406
Developmental Biology
13 Jan 2026
Pham, C.N., Castelli, F., Finet, F., Leroy, C., Chollet, C., Chirayath, T.W., Moitra, S., Zarka, M., Ostertag, A., Brial, F., Combes, C., Latourte, A., Bardin, T., Fenaille, F., Richette, P. and Ea, H.K. (2026), Spermidine Reproduces the Anti-Inflammatory Effects of Intermittent Fasting and Prevents Urate and Calcium Pyrophosphate Crystal-Induced Inflammation. Arthritis Rheumatol (2025)
Inflammation
18 Nov 2025
Wu, C., Wang, T., Ghosh, A. et al. MTCH2 modulates CPT1 activity to regulate lipid metabolism of adipocytes. Nat Commun 16, 8831 (2025).
Metabolism