The cost of RNA-sequencing (RNA-seq) ranges from approximately $36.9 to $173 for a single sample in an mRNA-seq experiment. Sequencing costs have dropped significantly thanks to the ‘multiplexing’ of hundreds of samples in one sequencing run. The most expensive step is now often the library preparation; however, 3’ mRNA-seq barcoding and pooling library preparation methods are reducing these costs dramatically (Figure 1). Despite this, the true cost still depends on your experimental question.
This question will determine the most appropriate type of RNA-seq to perform, such as short-read mRNA, total RNA, small RNA, or long-read sequencing experiments.
Your experiment will also determine the number and type of samples, the RNA extraction method, type of library preparation, choice of sequencing platform, sequencing depth, read length and the use of single or paired-end (PE) reads. Less sequencing depth, shorter read lengths and single-end reads all make RNA-seq cheaper but reduce coverage.
Additional factors to consider include data analysis, storage and hands-on-time.
Here, we breakdown these costs for you (Figure 1).
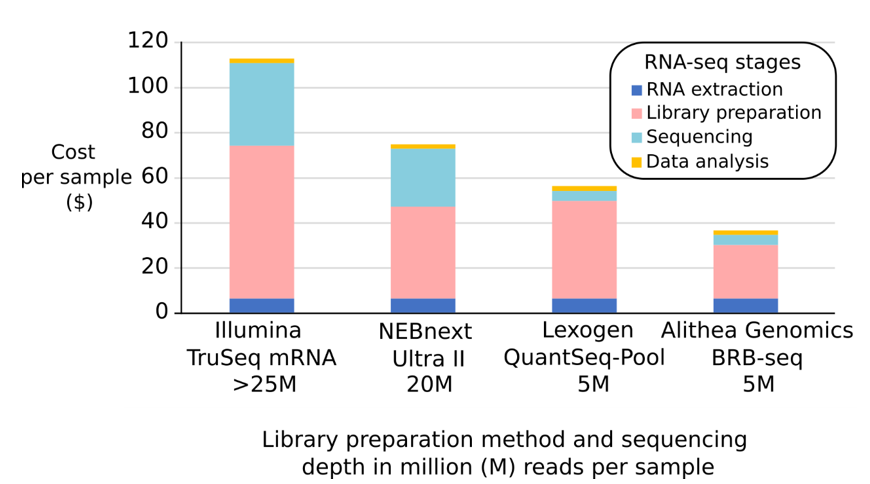
Figure 1: Breakdown of costs in mRNA-seq using different library preparation methods and the NovaSeq 6000 S4 300 cycle flow cell sequencing option from Illumina for 150bp PE-reads. Labor costs are excluded.
RNA extraction:
The first step is to extract high-quality RNA. For cell or tissue samples, labs commonly use solvent-based TRIzol ($2.2 per sample) [1] or silica-based column kits such as the QIAgen RNeasy Kit ($7.1 per sample) [2]. RNA quality is then checked with a Bioanalyzer RNA-6000-Nano chip ($4.1 per sample) [3].
Cost per sample: $6.3 to $11.2.
Library preparation:
The second step is to prepare a sequencing library for each sample of extracted RNA. This step is now often the most expensive part of an RNA-seq experiment (Figure 1).
For mRNA-seq, the cost of Illumina’s TruSeq mRNA stranded library prep kit, indexes and free-adapter blocking reagent is $64.4 per sample [4].
The NEBnext Ultra II RNA library prep kit for Illumina is $37 per sample [5], whilst the QuantSeq-Pool Sample-Barcoded 3' mRNA-Seq Library Prep Kit from Lexogen has a similar cost at $39.5 per sample [6].
Technologies such as Bulk RNA Barcoding and sequencing (BRB-seq) significantly reduce these costs by early barcoding and pooling of samples so only one subsequent library preparation is then required [7, 8]. This makes the Alithea Genomics MERCURIUS BRB-seq library preparation kit by far the most cost-effective option at $19.7 per sample [8].
Library quality can be assessed for all preparations using a Bioanalyzer DNA-1000 chip for $4.3 per sample [9].
Cost per sample (Illumina TruSeq): $68.7
Cost per sample (NEBnext Ultra II): $41.3
Cost per sample (Lexogen QuantSeq-Pool): $43.8
Cost per sample (Alithea MERCURIUS BRB-seq): $24
Sequencing:
The third step is to sequence the prepared libraries.
Instruments such as the Illumina NovaSeq 6000 have various flow cell options with different sequencing outputs; however, if a flow cell is not at full capacity, cost per sample can soar.
For example, on the NovaSeq 6000 instrument, the ‘SP’ 300 cycle flow cell is ideal for smaller experiments, as around 32 samples can be multiplexed to detect robustly expressed transcripts with ≥25M reads per sample when using the Illumina TruSeq library preparation [10]. It gives 1.3 to 1.6 billion 150bp PE reads (200 to 250Gb of data) for $96 per sample when using TruSeq if the flow cell is at full capacity [10, 11].
The NEBnext Ultra II library preparation allows a similar sensitivity at a reduced sequencing depth of 20M, allowing multiplexing of around 65 samples at a cost of $47.3 per sample.
Results comparable to Illumina TruSeq can also be achieved with a far lower sequencing depth of 5M reads by using 3’ mRNA-seq multiplexing library preparations such as Lexogen QuantSeq-Pool or Alithea Genomics MERCURIUS BRB-seq. This allows 260 samples to be multiplexed and results in a cost per sample of $11.8.
Cost per sample (Illumina TruSeq; ≥25M reads): $96
Cost per sample (NEBnext Ultra II; 20M reads): $47.3
Cost per sample (Lexogen QuantSeq-Pool; 5M reads): $11.8
Cost per sample (Alithea MERCURIUS BRB-seq; 5M reads): $11.8
The highest capacity ‘S4’ 300 cycle flow cell is the most cost-effective for high-throughput studies as approximately 400 samples can be multiplexed for ≥25M reads per sample [10]. It generates 16 to 20 billion 150bp PE reads (3000Gb of data) for $36.9 per sample when using Illumina TruSeq library preparation, if the flow cell is at full capacity [10, 11].
Sequencing of the NEBnext Ultra II library preparation costs $25.9 per sample, when using a sequencing depth of 20M on a NovaSeq 600 S4 flow cell.
The most cost-effective option is again the use of Lexogen QuantSeq-Pool or Alithea Genomics MERCURIUS BRB-seq, as a far lower sequencing depth of 5M reads is sufficient for accurate gene expression quantification comparable to TruSeq. An astonishing 3200 samples can be multiplexed on a NovaSeq 600 S4 flow cell for a cost of $4.6; a total cost similar to assaying four genes by qRT-PCR.
Cost per sample (Illumina TruSeq at ≥25M): $36.9
Cost per sample (NEBnext Ultra II; 20M reads): $25.9
Cost per sample (Lexogen QuantSeq-Pool; 5M reads): $4.6
Cost per sample (Alithea MERCURIUS BRB-seq; 5M reads): $4.6
Data analysis and storage:
Many free, open-source tools exist for post-sequencing analyses [12]. Mapping reads to a reference genome can be done using Bowtie2 [13] or STAR [14], whilst tools like DEseq [15] can be used for normalization and differential expression analyses. These are suitable for bioinformaticians, but biologist-friendly online tools such as Galaxy [16] require no bioinformatics experience.
Cloud-based analytic pipelines with integrated storage such as Illumina BaseSpace are available for around $2 per sample and 1000Gb storage at $22.5 per month [17]. A simple, one-click cloud-based pipeline for the analysis of BRB-seq is also available from Alithea Genomics and is free to use [18].
Cost per sample: approximately $2 (plus ongoing storage costs)
Hands-on-time:
Factor in approximately three- or four-days to complete all stages.
Overall estimated cost (on S4 flow cell at full capacity, excluding labor and instrument purchase):
Cost per sample (Illumina TruSeq at ≥25M): $113.9
Cost per sample (NEBnext Ultra II; 20M reads): $75.5
Cost per sample (Lexogen QuantSeq-Pool; 5M reads): $56.7
Cost per sample (Alithea MERCURIUS BRB-seq; 5M reads): $36.9
References:
[1] https://www.fishersci.com/shop/products/ambion-trizol-reagent-2/p-4918750
[2] https://www.qiagen.com/us/products/discovery-and-translational-research/dna-rna-purification/rna-purification/total-rna/rneasy-kits/
[3] https://www.fishersci.com/shop/products/rna-6000-nano-kit-5/NC1783726
[6] https://dmarkbio.com/products/lex-139
[7] Alpern, Daniel, et al. "BRB-seq: ultra-affordable high-throughput transcriptomics enabled by bulk RNA barcoding and sequencing." Genome biology 20.1 (2019): 1-15.
[8] https://www.alitheagenomics.com/mercurius-brb-seq-kit
[9] https://www.fishersci.com/shop/products/agilent-dna-1000-kit-3/NC1758992
[10] https://emea.illumina.com/systems/sequencing-platforms/novaseq/specifications.html
[11] https://www.illumina.com/products/by-type/sequencing-kits/cluster-gen-sequencing-reagents/novaseq-reagent-kits.html
[12] Conesa, Ana, et al. "A survey of best practices for RNA-seq data analysis." Genome biology 17.1 (2016): 1-19.
[13] Langmead, Ben, and Steven L. Salzberg. "Fast gapped-read alignment with Bowtie 2." Nature methods 9.4 (2012): 357-359.
[14] Dobin, Alexander, et al. "STAR: ultrafast universal RNA-seq aligner." Bioinformatics 29.1 (2013): 15-21.
[15] Anders, Simon, and Wolfgang Huber. "Differential expression analysis for sequence count data." Nature Precedings (2010): 1-1.
[16] Goecks, Jeremy, Anton Nekrutenko, and James Taylor. "Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences." Genome biology 11.8 (2010): 1-13.
[17] https://www.illumina.com/products/by-type/informatics-products/icredits.html
[18] https://www.alitheagenomics.com/software

.jpg?width=624&height=405&name=what-is-a-budget%20(2).jpg)


