RNA-seq quantifies the genome-wide level of thousands of mRNA transcripts from many samples in a single assay. Researchers can now choose from many next-generation RNA-seq library preparation protocols, and the recommended amount of RNA needed for each of these varies by platform.
Short-read RNA-seq approaches often recommend 10 ng to 1 µg of input RNA. However, the minimum requirement for long-read RNA-seq protocols is higher at 300 ng to 500 ng. For samples with RNA quantities as low as 100 pg, ultra-low-input sequencing options now provide transcriptomic data as reliable as standard RNA-seq protocols. Novel bulk 3’mRNA-seq technologies are now suitable for standard and low-input RNA quantities.
This article discusses how much RNA you will need for short or long-read RNA-seq experiments and explores novel options for ultra-low-input RNA-seq.
The importance of RNA quantity in RNA-seq
When you plan your RNA-seq experiment, it is essential to use the amount of RNA recommended by your desired RNA-seq protocol.
If too little RNA is used, transcriptomic data may be less accurate and less reliable and can limit novel findings (Bhargava et al., 2014; Shanker et al., 2015). This is due to an increased likelihood of lower RNA-seq library complexities and increased sequencing artifacts.
Read on to explore the recommended amounts of RNA for different RNA-seq library preparation protocols and novel bulk 3’mRNA-seq approaches. In practice, the amount of RNA required will often be more than the minimum recommended amount due to RNA quality control stages before library preparation.
RNA quantities for short-read RNA-seq approaches
Short-read RNA-seq relies on fragmenting transcripts into smaller 200 to 500 base pair pieces, followed by reassembly during data analysis.
For short-read sequencing approaches, the gold standard TruSeq Stranded mRNA Library Prep from Illumina requires a minimum of 100 ng of total RNA (Table 1) (Illumina, 2023).
Other library preparation methods require tenfold less total RNA than the TruSeq method. For example, NEBNext® Ultra™ II Directional RNA Library Prep Kit for Illumina® from New England Biolabs and the MGIEasy RNA Library Prep Set from MGI require only 10 ng of total RNA (Table 1) (MGI, 2023; New England Biolabs, 2023).
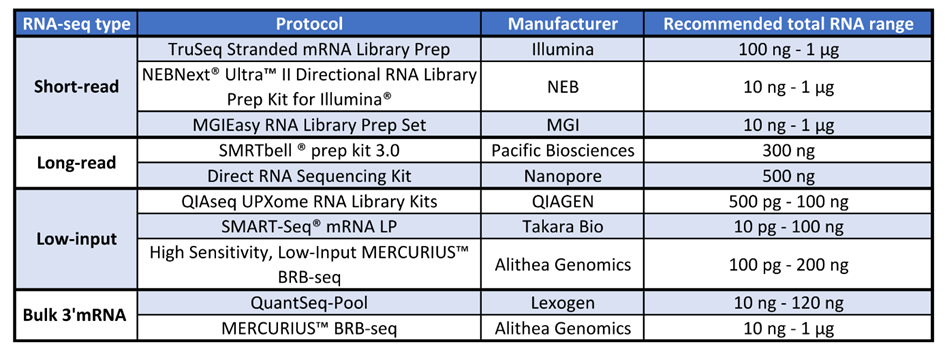
Table 1 – Table indicating the type of RNA-seq approach, examples of different protocols for each approach, the manufacturer, and the amount of total RNA recommended by each protocol.
RNA quantities for long-read RNA-seq approaches
Long-read RNA-seq technologies differ from short-read approaches. They sequence entire intact mRNA molecules. The Single Molecule Real-Time (SMRT) sequencing technology from Pacific Biosciences produces an average read length of 20 kb (Pacific Biosciences, 2023). In addition, Nanopore sequencing from Oxford Nanopore Technologies provides ultra-long-reads of an average of 100 kb (Oxford Nanopore Technologies, 2023).
Long-read RNA-seq approaches were named Method of the Year 2022 by Nature Methods thanks to their power to detect accurate transcript isoforms, splice variants, and fusion transcripts to provide a complete picture of the transcriptome (Nature Methods Editorial, 2023).
At present, long-read RNA-seq approaches require a greater quantity of total RNA for library preparation. For example, 300 ng of total RNA is needed for the SMRTbell ® prep kit 3.0 from Pacific Biosciences, and 500 ng is required for the Nanopore Direct RNA Sequencing Kit (Table 1) (Oxford Nanopore Technologies, 2023; Pacific Biosciences, 2023).
RNA quantities for low-input RNA-seq approaches
In some cases, RNA-seq libraries must be prepared from poor-quality or low-quantity samples such as formalin-fixed, paraffin-embedded samples, sorted cell subtypes, or tissue samples.
To overcome these challenges, several low-input RNA-seq library preparation technologies now enable robust transcriptomics, even from suboptimal RNA (Palomares et al., 2019).
The QIAseq UPXome RNA Library Kits from QIAGEN require 500 pg to 100 ng, whereas the SMART-Seq® mRNA LP from Takara Bio requires a minimum of 10 pg of total RNA (Table 1) (QIAGEN, 2023; Takara Bio, 2023).
A novel 3’mRNA-seq method for low amounts of RNA is the High Sensitivity, Low-Input MERCURIUS™ BRB-seq kit from Alithea Genomics. It requires a minimum of 100 pg of total RNA (Table 1) (Alithea Genomics, 2023a).
RNA quantities for bulk 3’mRNA-seq approaches
In contrast to short and long-read approaches, bulk 3’mRNA-seq technologies add a sample barcode to the 3’ end of mRNA transcripts (Alpern et al., 2019).
QuantSeq-Pool from Lexogen requires 10 ng to 120 ng of total RNA (Table 1) (Lexogen, 2023).
In contrast, MERCURIUS™ BRB-seq from Alithea Genomics allows a greater range of RNA inputs, from 10 ng to 1 μg of RNA (Table 1) (Alithea Genomics, 2023b).
Contact us at info@alitheagenomics.com for more information about how MERCURIUS™ BRB-seq can help your RNA-seq study.
References
- Alithea Genomics, 2023a. MERCURIUS BRB-seq kit for Illumina. Available at: https://alitheagenomics.com/products/mercurius-brb-seq-kit
- Alithea Genomics, 2023b. MERCURIUS High sensitivity low-input BRB-seq service. Available at: https://alitheagenomics.com/services/high-sensitivity-low-input-brb-seq
- Alpern, D., Gardeux, V., Russeil, J., Mangeat, B., Meireles-Filho, A.C., Breysse, R., Hacker, D. and Deplancke, B., 2019. BRB-seq: ultra-affordable high-throughput transcriptomics enabled by bulk RNA barcoding and sequencing. Genome biology, 20(1), pp.1-15.
- Bhargava, V., Head, S.R., Ordoukhanian, P., Mercola, M. and Subramaniam, S., 2014. Technical variations in low-input RNA-seq methodologies. Scientific reports, 4(1), pp.1-10.
- Illumina, 2023. TruSeq Stranded mRNA | Sequence mRNA samples. Available at: https://www.illumina.com/products/by-type/sequencing-kits/library-prep-kits/truseq-stranded-mrna.html
- Lexogen, 2023. QuantSeq 3’ mRNA-Seq Family. Available at: https://www.lexogen.com/quantseq-family/
- MGI, 2023. MGIEasy RNA Library Prep Set. Available at: https://en.mgi-tech.com/products/reagents_info/id/3
- Nature Methods Editorial, 2023. Method of the Year 2022: long-read sequencing. Nat Methods 20, 1.
- New England Biolabs, 2023. NEBNext Ultra II Directional RNA Library Prep Kit for Illumina | NEB. Available at: https://international.neb.com/products/e7760-nebnext-ultra-ii-directional-rna-library-prep-kit-for-illumina
- Oxford Nanopore Technologies, 2023. Direct RNA Sequencing Kit. Available at: https://nanoporetech.com
- Pacific Biosciences, 2023. Preparing Iso-Seq libraries using SMRTbell prep kit 3.0. Available at: https://www.pacb.com/wp-content/uploads/Procedure-checklist-Preparing-Iso-Seq-libraries-using-SMRTbell-prep-kit-3.0.pdf
- Palomares, M.A., Dalmasso, C., Bonnet, E., Derbois, C., Brohard-Julien, S., Ambroise, C., Battail, C., Deleuze, J.F. and Olaso, R., 2019. Systematic analysis of TruSeq, SMARTer and SMARTer Ultra-Low RNA-seq kits for standard, low and ultra-low quantity samples. Scientific Reports, 9(1), p.7550.
- QIAGEN, 2023. QIAseq UPXome RNA Library Kits. Available at: https://www.qiagen.com/de-us/products/discovery-and-translational-research/next-generation-sequencing/rna-sequencing/ultraplex/qiaseq-upxome-rna-library-kits?catno=334702
- Shanker, S., Paulson, A., Edenberg, H.J., Peak, A., Perera, A., Alekseyev, Y.O., Beckloff, N., Bivens, N.J., Donnelly, R., Gillaspy, A.F. and Grove, D., 2015. Evaluation of commercially available RNA amplification kits for RNA sequencing using very low input amounts of total RNA. Journal of biomolecular techniques: JBT, 26(1), p.4.
- Takara Bio, 2023. SMART-Seq mRNA LP and SMART-Seq mRNA. Available at: https://www.takarabio.com/products/next-generation-sequencing/rna-seq/ultra-low-input-rna-seq/smart-seq-mrna-lp-and-smart-seq-mrna




