RNA sequencing (RNA-seq) provides a molecular snapshot of the cellular transcriptome, offering a treasure trove of information regarding gene regulation and expression. One of the most common—and arguably the most biologically relevant—applications of RNA sequencing is the ability to assess differential gene expression (DGE), which provides invaluable data on genome-wide differences in gene expression between biological samples. As changes in gene expression often involve multiple comparisons—such as between wild types and mutants, life history stages, experimental time points, or environmental conditions—and normally include controls and replicates, DGE analyses often involve numerous RNA samples. However, until recently, traditional RNA-seq strategies have been too laborious, expensive, and analytically challenging to replace reverse transcription-quantitative real-time PCR (qRT-PCR) as the preferred method of quantifying changes in gene expression, thereby limiting the number of RNA samples that can be processed (Alpern et al. 2019).
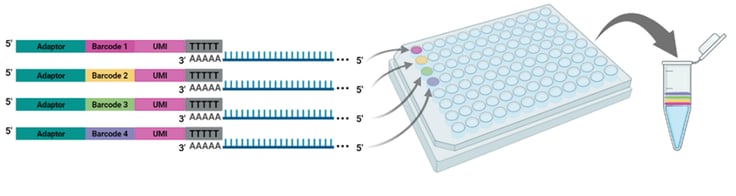
Figure 1. Each well of the BRB-seq oligo(dT) primer plate contains a distinct barcode adaptor to uniquely label the RNA sample added to each well. After reverse transcription, barcoded samples are pooled for the remainder of the cDNA library preparation workflow and library sequencing.
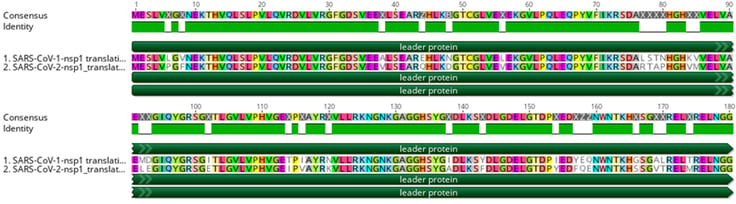
As incubation temperature has been previously shown to affect replication kinetics in coronaviruses and other respiratory viruses (Hoorn and Tyrrell 1966; Corman et al. 2016), a recent study by V’kovski et al. (2021) used BRB-seq to assess DGE between coronaviruses SARS-CoV-1 and SARS-CoV-2 at two biologically relevant temperatures (33 and 37°C) to mimic the upper and lower respiratory tracts, respectively. Despite their phylogenetic similarities, most notably in functionally conserved regions (Figure 2), SARS-CoV-1 and SARS-CoV-2 are known to exhibit considerable differences in human-to-human transmission dynamics, pathogenicity, and clinical courses of infection (Cevik, Kuppalli, et al. 2020; Cevik, Tate, et al. 2020), indicating possible differences in virus-host dynamics (V’kovski et al. 2021).
The authors found that while SARS-CoV-1 and SARS-CoV-2 replicated to similar titers over the course of infection at 37°C (i.e., lower airway), SARS-CoV-2 infection resulted in 10-fold higher titers at 33°C (i.e., upper airway) between 72 and 96 h post infection. Furthermore, viral replication was attenuated in both viruses after incubation with interferons at both experimental temperatures. Given the substantial effect of incubation temperature on replication kinetics in SARS-CoV-2, the authors sought to investigate the effects of the two experimental temperatures on the host transcriptional response to SARS-CoV-1 and SARS-CoV-2 at 24, 48, 72, and 96 h post infection using BRB-seq. Samples were sequenced at a depth of 12 million reads per sample to ensure that genes with lower levels of expression were captured in the analysis. BRB-seq allowed for the downstream analysis of three distinct approaches to DGE: i) a comparison of DGE between SARS-CoV-1 and SARS-CoV-2 in response to temperature, ii) pairwise comparisons between each virus and control cultures at each timepoint, and iii) an analysis of changes in gene expression over time.
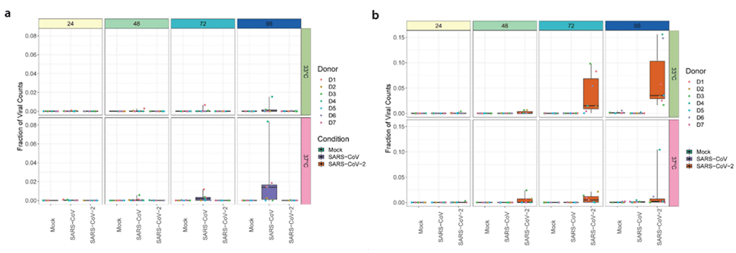
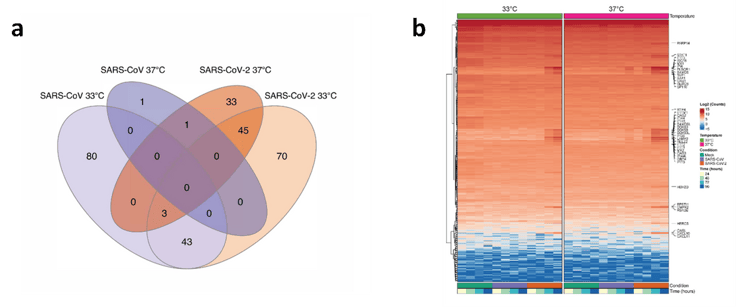
Figure 4. Differential gene expression as assessed using BRB-seq technology. a) Venn diagram illustrating the overlap in differentially expressed genes between SARS-CoV-1 and SARS-CoV-2 infected cultures at 33 and 37°C. b) Hierarchical cluster analysis of DEGs identified in SARS-CoV-1 and SARS-CoV-2 infected cultures compared to control (i.e., mock) cultures at 33 or 37°C. Figure adapted from V’kovski et al. (2021).
In conclusion, both the increased replication and increased quantity of sequence reads for SARS-CoV-2 at an incubation temperature that mimics the upper respiratory tract supports real-world observations of high levels of SARS-CoV-2 replication in the upper airway, as well as increased transmissibility in comparison to SARS-CoV-1. In this study, BRB-seq facilitated complex transcriptomic comparisons between multiple RNA samples, thereby providing a roadmap to better understanding the virus-host dynamics of the pandemic virus SARS-CoV-2.
For more information about BRB-seq and its application to COVID-19 and research on infectious diseases, do not hesitate to contact us at info@alitheagenomics.com
References
- Alpern, Daniel, Vincent Gardeux, Julie Russeil, Bastien Mangeat, Antonio C. A. Meireles-Filho, Romane Breysse, David Hacker, and Bart Deplancke. 2019. “BRB-Seq: Ultra-Affordable High-Throughput Transcriptomics Enabled by Bulk RNA Barcoding and Sequencing.” Genome Biology 20 (1): 71. https://doi.org/10.1186/s13059-019-1671-x.
- Cevik, Muge, Krutika Kuppalli, Jason Kindrachuk, and Malik Peiris. 2020. “Virology, Transmission, and Pathogenesis of SARS-CoV-2.” BMJ, October, m3862. https://doi.org/10.1136/bmj.m3862.
- Cevik, Muge, Matthew Tate, Ollie Lloyd, Alberto Enrico Maraolo, Jenna Schafers, and Antonia Ho. 2020. “SARS-CoV-2, SARS-CoV-1 and MERS-CoV Viral Load Dynamics, Duration of Viral Shedding and Infectiousness – a Living Systematic Review and Meta-Analysi.” https://doi.org/10.1101/2020.07.25.20162107.
- Corman, Victor M., Isabella Eckerle, Ziad A. Memish, Anne M. Liljander, Ronald Dijkman, Hulda Jonsdottir, Kisi J. Z. Juma Ngeiywa, et al. 2016. “Link of a Ubiquitous Human Coronavirus to Dromedary Camels.” Proceedings of the National Academy of Sciences 113 (35): 9864–69. https://doi.org/10.1073/pnas.1604472113.
- Hoorn, B., and D. A. J. Tyrrell. 1966. “A New Virus Cultivated Only in Organ Cultures of Human Ciliated Epithelium.” Archiv Für Die Gesamte Virusforschung 18 (2): 210–25. https://doi.org/10.1007/BF01241842.
- V’kovski, Philip, Mitra Gultom, Jenna N. Kelly, Silvio Steiner, Julie Russeil, Bastien Mangeat, Elisa Cora, et al. 2021. “Disparate Temperature-Dependent Virus–Host Dynamics for SARS-CoV-2 and SARS-CoV in the Human Respiratory Epithelium.” PLOS Biology 19 (3): e3001158. https://doi.org/10.1371/journal.pbio.3001158.



