Immunotherapy itself is an approach that aims at using or re-purposing a patient's immune system to fight back cancer.
As for all therapies - and in particular the ones targeting complex diseases such as cancer - the efficacy of immunotherapy can greatly be improved by personalizing and tailoring treatments to patients. However, this is unfortunately easier said than done!
In fact, the scientific and medical community still lack reliable biomarkers and interpretation tools to be able to reliably personalize immunotherapy.
Luckily, accumulating evidence and emerging tools are opening exciting opportunities.
In this context, both bulk and single-cell RNA sequencing (RNA-seq) constitute great resources for the development of new knowledge and new biomarkers that can help provide the right therapy at the right time (Fig. 1).
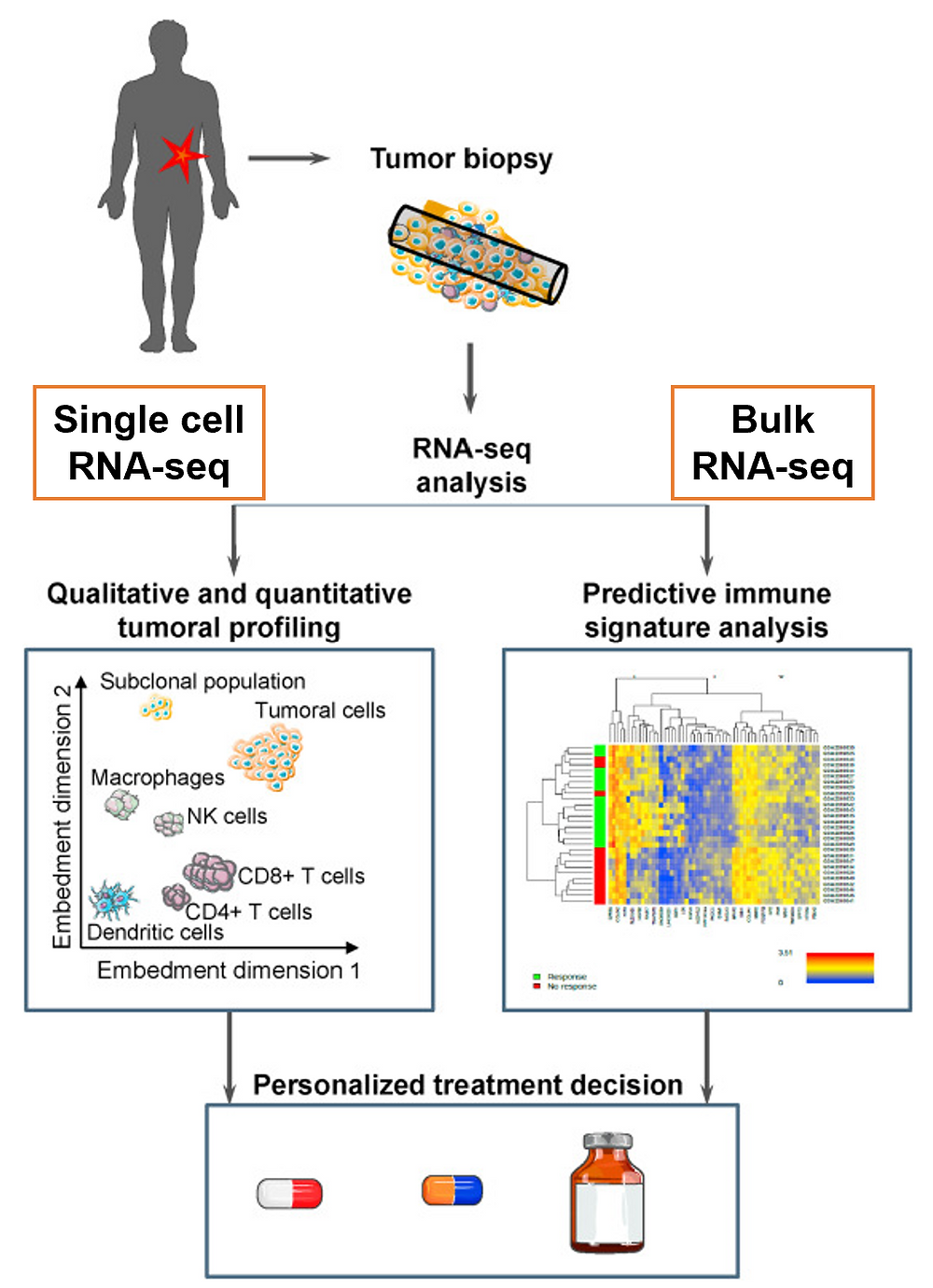
Figure 1 (adapted from the article)
· NCT03739372, which aims to guide treatment for children with high-grade gliomas.
· NCT04318834, which examines a cohort of adult patients with biliary tract cancers.
· NCT03784014 and NCT03375437, which study soft-tissue sarcomas.
Finally, another interesting class of clinical studies used RNA-seq to evaluate the response of tumours after receiving a type of therapeutic agent called immune-checkpoint inhibitors (NCT03978624, NCT04326257 and NCT03673787).
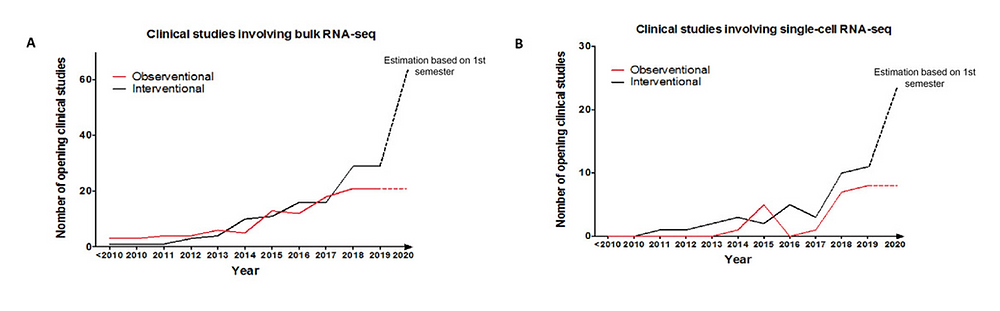
However, we believe the clinical translation of RNA-seq will require further efforts specifically aimed the generation of more diverse and well-curated data, for example from international consortia.
Finally, defining standardised analysis pipelines among thousands available, from the wet to dry lab, will be essential and will greatly benefit the reproducibility and gradual generalization of the approach.
Please contact us if you have more questions about how bulk and single-cell RNA-seq can help the immunology research and the discovery of new biomarkers and therapies.



