Probe-based gene expression profiling methods, such as Affymetrix™ GeneChip™ microarrays, QuantiGene™ Plex assays from Invitrogen™, and more recently the Clariom™ GO Screen, were a mainstay of toxicogenomic studies for drug discovery and development in previous decades thanks to their customization and automation options.
But, probe-based technologies limit the researcher to detecting transcripts that correspond to existing genomic sequence information and generally have low scalability, driving a relatively high cost per sample for limited transcriptomic read-outs.
The emergence of RNA-seq-based methods has helped overcome these challenges by allowing researchers to investigate both known transcripts and explore new ones. Continued innovation has also recently propelled us into the ultra-scalable transcriptomic profiling era. Now, the development of bulk 3’ mRNA-seq technologies like MERCURIUS™ DRUG-seq offers unbiased transcriptome-wide profiling for hundreds of samples simultaneously, with no prior probe design, at an ever-decreasing cost per sample.
In this article, we compare probe-based gene expression profiling methods to ultra-high-throughput MERCURIUS™ DRUG-seq from Alithea Genomics so you can evaluate which may be most appropriate for integration with your toxicogenomic, drug discovery, or development pipeline.
Targeted transcript detection versus unbiased 3’ transcript detection
Before commencing any gene expression profiling study, it’s essential to understand each technology's capabilities, from the number and type of transcripts detected, to sample requirements and throughput, to ensure robust and informative transcriptomic data to inform critical decision points in a project's pipeline.
Probe-based gene expression profiling technologies rely on the stringent hybridization of pre-defined oligonucleotide sequences to desired transcripts of interest but cannot detect novel transcripts, whereas bulk 3’ mRNA-seq methods provide a more unbiased view of the whole transcriptome as unique barcodes label each mRNA transcript in a sample.
The Clariom™ GO Screen and Affymetrix™ GeneChip™ Human Transcriptome Array 2.0 are both microarray probe-based approaches (Table 1) (Thermo Fisher Scientific, 2023a, b). The Clariom™ GO Screen generates expression read-outs for around 20,000 protein-coding genes, while the GeneChip™ array provides a more comprehensive expression profile, including 45,000 protein-coding genes, and around 40,000 non-coding transcripts (Table 1). Over 300,000 other probe sets span exon-exon junctions to provide alternative splicing information
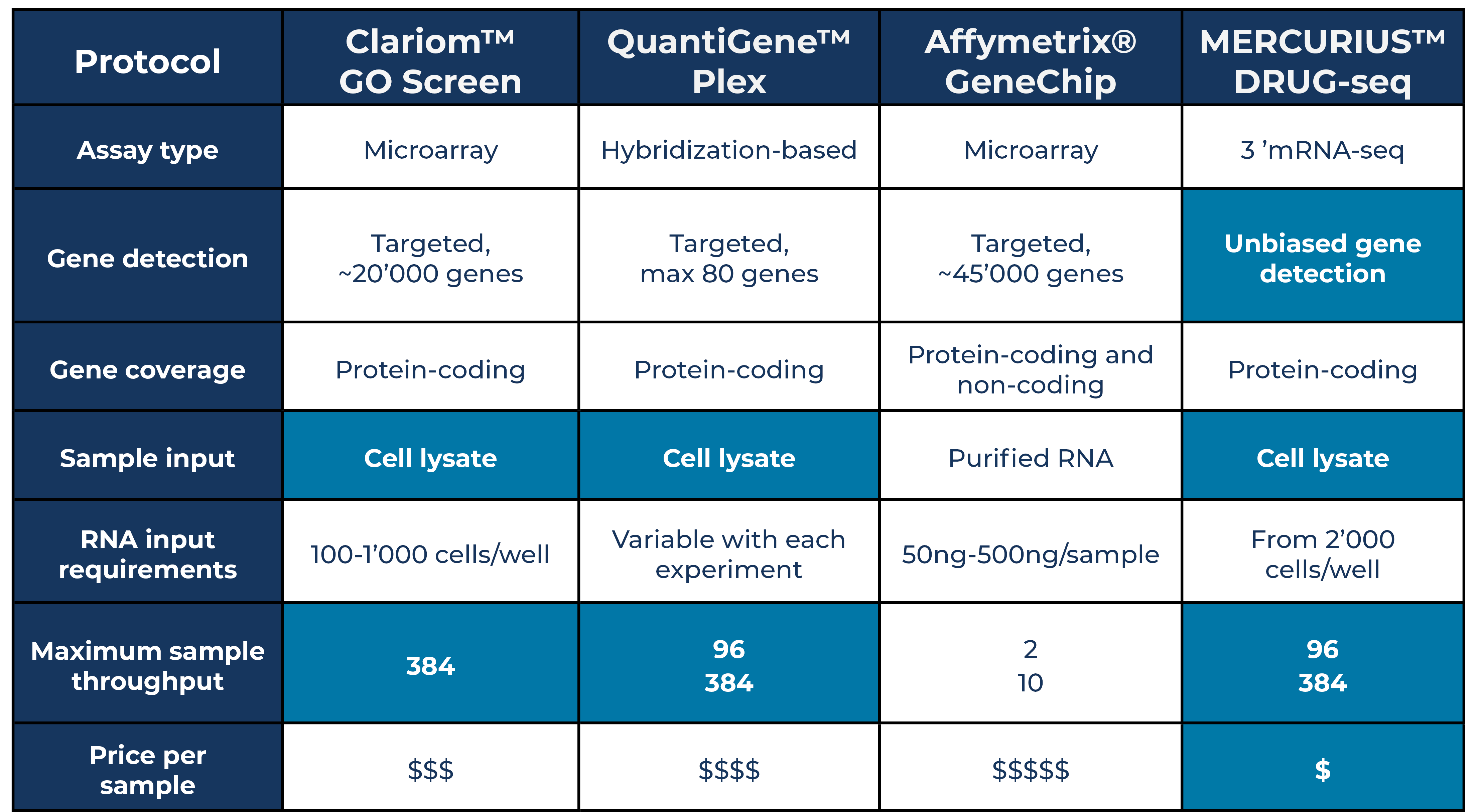
Table 1. Table comparing different features of probe-based and 3’ mRNA-seq technologies.
In contrast, another probe-based technology called the QuantiGene™ Plex assay uses branched DNA technology combined with Luminex™ xMAP™ beads to provide a more restricted view of the transcriptome limited to a maximum of 80 transcripts, suitable for known biomarker-detection studies (Table 1) (Thermo Fisher Scientific, 2023c).
For unbiased detection of protein-coding transcriptome-wide expression levels not reliant on known sequence information, the 3’ mRNA-seq-based method MERCURIUS™ DRUG-seq provides expression read-outs for around 20,000 genes at a shallow sequencing depth of five million reads per sample (Table 1). Unlike target technologies, users can opt for deeper sequencing to enable the detection of more lowly expressed genes if a study requires this.
Also, novel transcripts are readily detected due to capture with highly optimized barcoded oligo(dT) primers that uniquely tag each individual mRNA molecule and sample during the first-strand synthesis step of cDNA library preparation (Alithea Genomics, 2023).
Although bulk 3’ mRNA-seq technologies do not allow for the analysis of full-length transcripts, splice variants, or fusion genes, generating a sequencing library of complete transcripts is arguably unnecessary when a broad overview of protein-coding gene expression is required.
GeneChip™, QuantiGene™, Clariom™ GO, and MERCURIUS™ DRUG-seq sample requirements
For large-scale compound screens requiring a transcriptomic read-out, sample compatibility with the chosen gene expression profiling technology is also critical to generating accurate and reliable data to evaluate toxicity, detect biomarkers, or establish a mechanism of action. Whether RNA must be extracted before transcriptomic analysis is also crucial, as it will increase both cost and hands-on time.
Affymetrix™ GeneChip™ arrays are not “plug and play” as they require input amounts of 500pg-10ng RNA per sample, which must be isolated before use, whether from whole blood, fresh/frozen tissue samples, or formalin-fixed paraffin-embedded (FFPE) tissue (Table 1).
Conversely, the Clariom™ GO Screen, QuantiGene™ Plex, and MERCURIUS™ DRUG-seq are optimized for use with diverse sample types directly on cell lysates with no prior RNA extraction required (Table 1).
The Clariom™ GO Screen requires 100-1,000 cells/well, variable amounts of cells are required for QuantiGene™ Plex assays depending on the experiment performed, and MERCURIUS™ DRUG-seq requires upwards of 2,000 cells/well (Table 1).
This ease of use makes these technologies suitable for automation and simplified integration into drug discovery and pharmacotranscriptomic pipelines.
Sample multiplexing and overall cost
Each technology's multiplexing capabilities and the cost per sample for gene expression profiling could also be a deal breaker for extensive studies with many samples.
Affymetrix™ GeneChip™ arrays have limited sample multiplexing options, with one array costing around $343.5, not including RNA extraction, making it the most expensive option per sample (Thermo Fisher Scientific 2023d).
QuantiGene™ Plex can be performed on plates containing isolated RNA from either 96 or 384 samples. The 384-well kit costs around $13 per sample but must be bought alongside QuantiGene™ Plex probe panels and extracted RNA kits at an additional cost (Thermo Fisher Scientific, 2023e).
The Clariom™ GO Screen is available for samples in 384-well plate format but is offered as a service only, making it the third most expensive option.
In contrast, MERCURIUS™ DRUG-seq is optimized for either 96 or 384-well plates but is further streamlined as all samples are multiplexed in one tube early in the workflow, providing a seamless, efficient library preparation process at a cost as low as $2 per sample (Alithea Genomics, 2023). A MERCURIUS™ DRUG-seq service is also offered at an additional cost.
Ultimately, the choice of either a probe-based gene expression profiling technology or a bulk 3’ mRNA-seq-based technology like MERCURIUS™ DRUG-seq will depend on different factors such as the aim of your study, your number of samples, the type of transcriptomic data required (alternative splicing detection, novel transcript detection, select biomarker detection), and the study budget.
To find out more about MERCURIUS™ DRUG-seq, please contact us by clicking here.
References
- MERCURIUS™ DRUG-seq Kits | Alithea Genomics (2023). Available at: https://alitheagenomics.com/products/drug-seq-kits
- GeneChip™ Human Transcriptome Array 2.0 Datasheet | Thermo Fisher Scientific (2023a). Available at: https://assets.thermofisher.com/TFS-Assets/LSG/brochures/hta_array_2_0_datasheet.pdf
- Clariom™ GO Screen Assay | Thermo Fisher Scientific (2023b). Available at: https://www.thermofisher.com/order/catalog/product/952361.
- QuantiGene™ Plex Gene Expression Assay Product Page | Thermo Fisher Scientific (2023c). Available at: https://www.thermofisher.com/us/en/home/life-science/gene-expression-analysis-genotyping/quantigene-rna-assays/quantigene-plex-assay.html
- GeneChip™ Human Transcriptome Array 2.0 Order Page | Thermo Fisher Scientific (2023d). Available at: https://www.thermofisher.com/order/catalog/product/902162
- QuantiGene™ Plex Gene Expression Assay Order Page | Thermo Fisher Scientific (2023e). Available at: https://www.thermofisher.com/order/catalog/product/QP1016




