Understanding the transcriptomic impacts of new compounds on cells is essential in drug development and toxicology. This process helps researchers evaluate a drug's on- or off-target effects, its toxicity, and potential mechanisms of action. Such insights are crucial for determining the safety and effectiveness of new therapeutic candidates.
To accelerate these discoveries, transcriptome profiling methods are rapidly evolving, focusing on increasing sample throughput and reducing costs. For compound screening and toxicology, this translates to larger screens with more experimental conditions and replicates, providing a wealth of information for critical decision-making.
However, keeping up with the fast-paced advancements in transcriptomic technologies can be challenging. In this blog post, we will compare two high-throughput transcriptome profiling technologies: MERCURIUS™ DRUG-seq from Alithea Genomics and TempO-Seq™ from BioSpyder™. We will examine how these technologies differ in terms of cost, scalability, and bias, helping you decide which method is best suited for your large-scale therapeutic or toxicology compound screens.
Transcriptome profiling for compound screening
RNA-seq for compound screening has traditionally involved costly and labor-intensive steps, including isolating and fragmenting mRNA, reverse transcribing mRNA to cDNA, and performing adapter ligation and PCR amplification. Each sample requires individual preparation, which increases both the cost and handling time, thereby limiting the throughput of samples.
Innovative library preparation methods like MERCURIUS™ DRUG-seq and TempO-Seq™ have emerged to overcome these limitations. These techniques utilize sample barcoding to create a single sequencing library containing multiple distinct samples, each employing unique strategies to achieve this efficiency.
Multiplexed technologies suitable for compound screening
TempO-Seq™ from BioSpyder™
This technology uses two detector oligos designed to hybridize adjacent target sequences in a given transcript (Yeakley et al., 2017). When both oligos are properly hybridized to the correct target locations, the two detector oligos are ligated, Excess unhybridized oligos are removed, and the ligated oligo pairs are amplified to add sample-specific barcodes and sequencing adapters. Sample barcodes permit the pooling of all samples before the seqencing (Fig.1A).
MERCURIUS™ DRUG-seq from Alithea Genomics
MERCURIUS™ DRUG-seq is a transformative tool for compound screening and drug discovery, combining unbiased, high-throughput compound screening with massively parallel and extraction-free transcriptomics. This method uses highly optimized and rigorously evaluated sample barcodes and unique molecular identifiers to tag the 3’ poly(A) tail of all mRNA molecules in a sample-specific manner during the first-strand synthesis step of cDNA library preparation (Fig.1B).
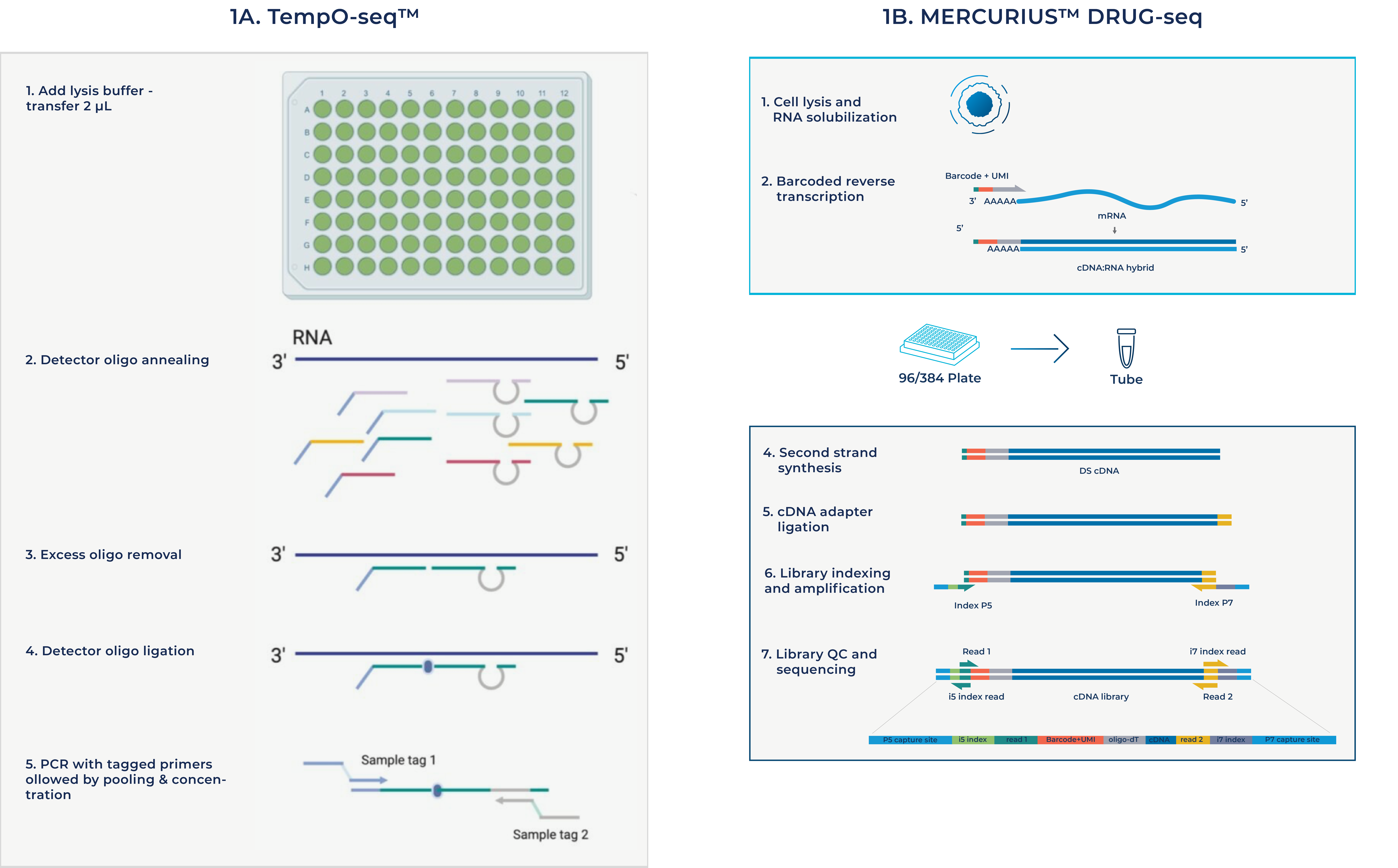
Figure 1 – The experimental workflows at a glance for (1A) TempO-Seq™ and MERCURIUS™ DRUG-seq (1B). Figure 1A is adapted from TempO-Seq™ | BioSpyder™ available at: https://www.biospyder.com/technology.
RNA extraction-free library prep
One key consideration for compound screening studies aiming to increase throughput while reducing cost and handling time is whether RNA must be extracted before library preparation.
TempO-Seq™ offers flexible inputs with purified RNA, cell lysates, or formalin-fixed paraffin-embedded tissues (FFPE) (Trejo et al., 2019).
The DRUG-seq protocol is designed to work with frozen cells and bypass RNA extraction. Thanks to the highly optimized lysis buffers for cell lysis of 2D cell cultures and organoid models, it efficiently generates library preps without prior RNA isolation.
The power of sample multiplexing
TempO-Seq™ allows the multiplexing of 96, 384, or 1,536 samples in the same tube, while it requires as little as 10 pg of RNA (the amount from a single cell) and provides insight into 20,000 pre-defined transcript sequences. This technology relies on late multiplexing – just before the sequencing step, which necessitates the processing of samples on a one-by-one basis.
The MERCURIUS™ DRUG-seq protocol allows researchers to multiplex up to 384 samples in one tube early on in the workflow. This enables scientists to process thousands of samples by combining the sample barcode indexing with the library indexing, which makes the design of the project very flexible. Moreover, it simplifies a lot the handling of the samples, which not only reduces the hands-on time but also reduces the cost significantly without compromising data quality. More than 15’000 genes can be detected at a seqencingg depth of 1.5M reads per sample.
Cost-effective compound screening
TempO-seq™ generally requires a probe set ranging from approximately 3’000 probes, optimized to target key pathways and correlated expression modules, to around 20’000 probes, enabling broad coverage of most known coding genes in the human genome (Mav et al., 2018).
Given the extensive probe design involved, the setup costs are likely very high. Besides the probe-set design, TempO-seq™ does not offer early sample multiplexing which means that all the enzymatic reactions and liquid handling need to be performed individually on every sample, from the beginning to the end of the workflow.
In contrast, the MERCURIUS™ DRUG-seq technology is based on early sample multiplexing to produce 3′ cDNA libraries, which provides great capacity for transforming large sets of samples into a unique sequencing library by targeting (without bias) the polyA tail of all mRNA transcripts. This provides seamless, efficient library preparation, offering significantly lower setup and running costs.
Download MERCURIUS™ DRUG-seq Pricing Guide
DRUG-seq technology: an unbiased approach
Another important aspect when performing compound screens is the use of an unbiased dataset.
The DRUG-seq technology provides an unbiased window into the transcriptome as it is not reliant on probes designed for known transcripts. This means it is highly amenable to discovering novel transcripts, a layer of information that may be missed with hybridization-based approaches such as TempO-Seq™. Additionally, its reliance on predefined gene content necessitates significant effort in template creation and meticulous probe sequence selection.
By capturing the entire transcriptome without bias, researchers can gain a comprehensive understanding of gene expression changes, identify unexpected transcriptomic signatures, and uncover novel pathways and biomarkers. This unbiased approach is crucial for revealing the full spectrum of biological effects induced by compounds.
|
Protocol |
TempO-seq™ |
MERCURIUS™ DRUG-seq |
|
Assay type |
Probe-based RNA-seq |
3’ mRNA-seq |
|
Gene detection |
Targeted, up to 20’000 genes |
Unbiased gene detection |
|
Genes profiled |
Protein-coding and non-coding |
Protein-coding |
|
Sample input |
Cell lysate |
Cell lysate |
|
RNA input requirements |
10pg or 1 cell/well |
2’000 cells per well |
|
Early sample multiplexing |
N/A |
Up to 384
|
|
Commercialized as |
Services |
Kits Services |
Request a Quote for the MERCURIUS™ DRUG-seq Kits
Conclusion
In conclusion, the significance of scalability, cost, and an unbiased RNA-seq outcome in high-throughput compound screening cannot be overstated. Cost-effective screening methods enable broader and more efficient testing, with many more samples and conditions to be tested. Meanwhile, an unbiased dataset ensures the reliability and accuracy of compound toxicity predictions, for example, minimizing the risk of adverse effects in later stages of drug development. Together, these factors are crucial for advancing safe and efficient compound screening.
Choosing the right technology for your compound screening needs can make a significant difference in the efficiency and success of your compound screening and drug discovery efforts. While TempO-Seq™ provides a sensitive and robust screening solution, its widespread adoption may be limited by the inherent cost and probe-based approach. On the other hand DRUG-seq offers a scalable, ultra-low cost and comprehensive molecular phenotyping assay, which may be better suited for truly large and hypothesis-free screening efforts.
References
- Yeakley, J.M. et al. (2017) ‘A trichostatin A expression signature identified by TempO-Seq targeted whole transcriptome profiling’, PloS one, 12(5), p.e0178302. Available at: https://doi.org/10.1371/journal.pone.0178302.
- Trejo, C.L. et al. (2019) ‘Extraction-free whole transcriptome gene expression analysis of FFPE sections and histology-directed subareas of tissue’, PLoS One, 14(2), p.e0212031. Available at: https://doi.org/10.1371/journal.pone.0212031.
- Mav D, Shah RR, Howard BE, et al. A hybrid gene selection approach to create the S1500+ targeted gene sets for use in high-throughput transcriptomics. PLOS ONE. 2018;13:e0191105. doi: 10.1371/journal.pone.0191105.