Ultra-low input RNA-seq library preparation methods like our MERCURIUS™ High Sensitivity BRB-seq kit provide researchers with powerful tools to explore the transcriptomes of small cell populations or precious clinical samples where extracted RNA quantities are minimal.
Most RNA-seq library preparation methods are optimized for a range of RNA quantities, so using the most appropriate technique for your particular samples will ensure the unbiased detection of the maximum number of genes possible. This is crucial for robust, reliable, and biologically meaningful discoveries, including novel disease biomarkers, cell marker genes, or hidden therapeutic targets.
But, at what RNA quantities should researchers consider using our low-input, high-sensitivity approach instead of standard MERCURIUS™ BRB-seq, and how much of a difference does it really make?
Read on to find out.
RNA Recommendations for Standard and High-Sensitivity BRB-seq
To generate the most optimal sequencing data with each technology, we suggest that all isolated total RNA samples have a minimum RNA integrity number (RIN) of 6 and an A260/230 ratio above 1.5 when quantifying by Nanodrop.
While RNA quality recommendations are the same for both technologies, the range of optimal RNA quantities differs between standard and high-sensitivity approaches.
We recommend 10 ng to 1 μg of purified RNA per well for standard BRB-seq, with a minimum of 1 μg total RNA per pool, and 1 ng to 500 ng of purified RNA per well for High Sensitivity BRB-seq, with a minimum of 96 ng total RNA per pool.
The correct choice of technique will also ultimately depend on the number of samples being pooled in your experiment and the total amount of RNA present in the pool (Fig. 1).
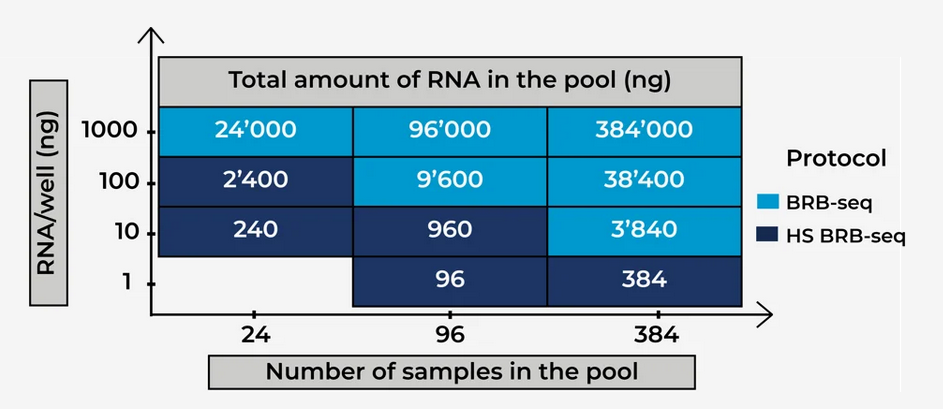
Figure 1. The choice between standard and High Sensitivity BRB-seq depends on the total amount of RNA in the pool.
Typically, where fewer samples are pooled, more RNA per sample is required before using standard BRB-seq, owing to the potential for reduced sequencing quality with lower-complexity reads from pools with low total RNA quantities.
For example, if pooling 24 RNA samples, each at 100 ng for a total pool RNA quantity of 2,400 ng, we still recommend using the high-sensitivity BRB-seq kit for optimal results.
When more samples are pooled, such as with the 384-sample formats, per sample RNA quantity can be as low as 10 ng for a total pool of 3,840 ng, but in each instance, we still suggest using as much RNA per sample as possible as per our recommendations above.
The more, the better.
If more RNA per sample isn’t feasible, robust, reproducible, and reliable sequencing data can still be obtained with the high-sensitivity BRB-seq kits for 96 and 384 samples with pooled RNA quantities as low as 96 ng or 384 ng at 1 ng of RNA per sample.
Also, it’s important to note that with both techniques, the RNA quantity, RIN, and 260/230 values of the starting RNA samples in the 24, 96, or 384 sample pools must be as uniform as possible, with a maximum of 10% variation to ensure an even distribution of reads after sequencing. If you’re unsure that your pool meets these criteria, please get in touch with us to discuss our shallow sequencing options for peace of mind that the library is of sufficient complexity to warrant deeper sequencing.
High Sensitivity BRB-seq Excels at Low RNA Inputs
Low amounts of RNA are typical for sorted cell subtypes, non-invasive tissue biopsies, or large-scale clinical or drug discovery studies. Using standard BRB-seq for these low abundance samples may reduce experimental efficiency, leading to missed genes potentially crucial as disease biomarkers, novel drug pathways, or core players in cellular functions.
So, to determine the relative detection sensitivities of each technology, we performed comparative experiments where we isolated RNA from 96 samples.
Different quantities of the same isolated RNA samples were pooled and prepared with standard and high-sensitivity BRB-seq, sequenced at 2 million reads per sample, and all data was analyzed with the same pipeline.
With 1 μg and 100 ng of RNA input, both standard and High Sensitivity BRB-seq detected around 20,000 expressed genes, with marginally more genes detected with standard BRB-seq (Fig. 2).
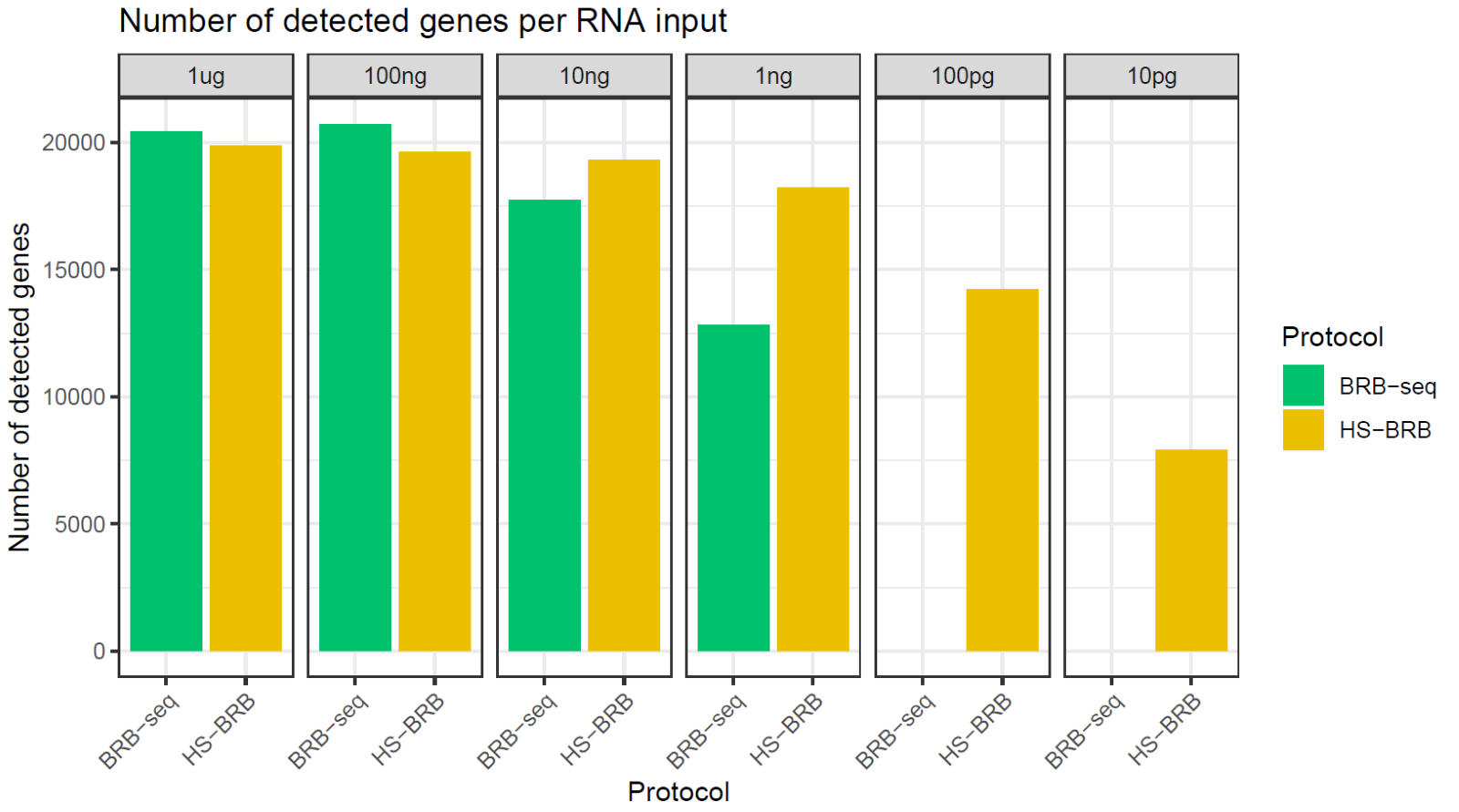
Figure 2. High Sensitivity BRB-seq excels with ultra-low-inputs of RNA. Bar chart comparing the number of genes detected by standard BRB-seq and High Sensitivity BRB-seq (HS-BRB) at different RNA input amounts. 96 samples were pooled together and sequenced at 2M reads/sample. All RNA was from the same samples.
At an input of 10 ng, High Sensitivity BRB-seq detected around 19,000 genes, 1,500 more than with standard BRB-seq.
But, at lower RNA inputs, High Sensitivity BRB-seq excelled.
The increased detection power of high-sensitivity BRB-seq was striking at 1 ng, where high-sensitivity BRB-seq detected around 18,000 genes, approximately 5,000 more than standard BRB-seq (Fig. 2).
When we dropped the input RNA quantity even lower to 100 pg, High Sensitivity BRB-seq detected around 14,000 expressed genes, whereas standard BRB-seq detected no genes (Fig. 2).
Similarly, at an ultra-low input of 10 pg, an RNA quantity equivalent to the total RNA in a single cell, High Sensitivity BRB-seq still detected around 8,000 expressed genes, all missed with standard BRB-seq (Fig. 2).
While these experiments explore the limits of our standard and High Sensitivity BRB-seq technologies and show that High Sensitivity BRB-seq is a powerful technology for the unbiased detection of gene expression at ultra-low-inputs, for most applications, we still suggest users use our minimum RNA recommendations to ensure the most robust and reproducible results possible.
For more information about how our MERCURIUS™ BRB-seq technologies can be used in your next low-input, high-throughput transcriptomic study, please get in touch with us.




