MERCURIUS™ BRB-seq and DRUG-seq library preparations are now compatible with MGI sequencing. This gives researchers maximum flexibility to perform ultra-high-throughput 3’ mRNA-seq experiments where MGI sequencers are available.
But how did we make the BRB-seq and DRUG-seq library prep workflow compatible with the sequencing technology from MGI?
Read on to find out.
What are BRB-seq and DRUG-seq?
MERCURIUS™ BRB-seq and DRUG-seq are robust, cost-effective, and ultra-scalable 3’ mRNA-seq library preparation technologies.
MERCURIUS™ DRUG-seq uses similar technology to BRB-seq but allows RNA extraction-free library preparation. It removes the costly and time-consuming RNA extraction step for large-scale transcriptomic screens with thousands of samples, such as in toxicology studies, as it generates library preps directly from cell lysates.
How do BRB-seq and DRUG-seq library preps work?
Both methods use optimized barcoded oligo(dT) primers, including unique molecular identifiers (UMIs), to uniquely tag the 3’ end of each mRNA molecule from each sample (Alpern et al., 2019). This occurs in the reverse transcription step of the cDNA library preparation (Fig. 1).
Thanks to these barcodes, researchers can multiplex hundreds of samples in the same tube and process them simultaneously from this point on.
Second-strand synthesis then creates barcoded full-length double-stranded cDNAs that are purified and fragmented (Fig. 1).
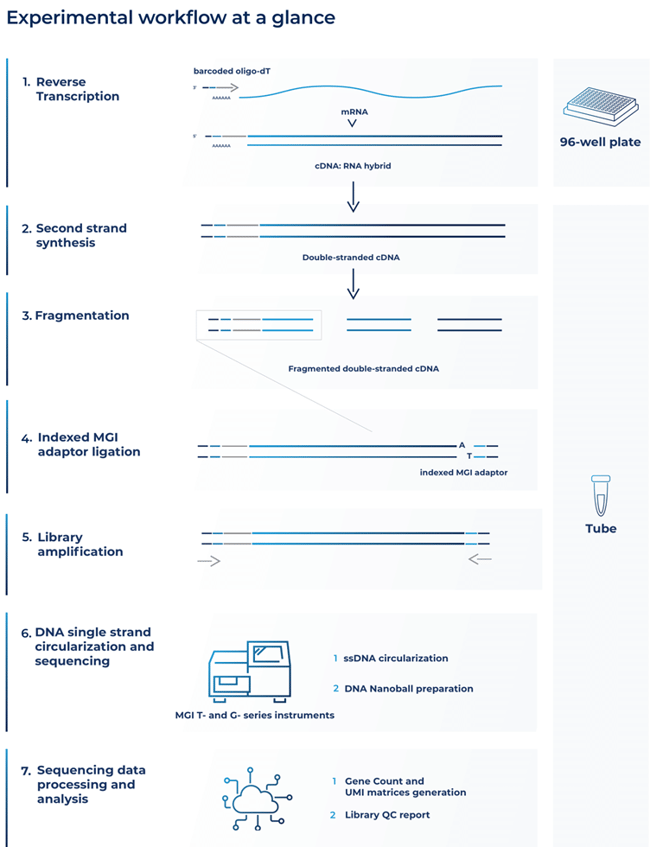
Figure 1. Diagram of the BRB-seq and DRUG-seq library prep workflow for MGI sequencing
How are MERCURIUS™ BRB-seq and DRUG-seq compatible with MGI sequencing?
The next stage in the library prep workflow is called index adapter ligation (Fig. 1). This stage makes BRB-seq and DRUG-seq compatible with MGI sequencing.
MERCURIUS™ BRB-seq and DRUG-seq kits for MGI seamlessly incorporate MGI library indexing adapters in the library prep workflow. Our kits include eight MGI library indexing adapters.
These adapters each contain a unique 10-nucleotide index sequence to allow the simultaneous sequencing of up to eight BRB-seq or DRUG-seq libraries in the same sequencing run.
Libraries prepared with different indexing pools can be sequenced on the same lane of the MGI sequencer.
Importantly, these index adapters also include all the necessary sequences for the binding of key oligos essential for library circularization and DNA nanoball preparation. These are crucial stages in the MGI library preparation process.
These index adapters allow researchers to make optimal use of the sequencing machine’s capacity thanks to ultra-high-throughput multiplexed experiments with thousands of samples.
After MGI index adapters are ligated, 5’ terminal fragments are amplified, size selected, and purified for fragments in the range of 200 to 800 base pairs with adapters on both sides (Fig. 1).
Barcoded, fully MGI-indexed double-stranded cDNAs of the correct fragment size are now ready for the MGIEasy circularization kit.
MGIEasy circularization after BRB-seq and DRUG-seq library prep
MGI sequencing uses an innovative approach based on the circularization of single-stranded DNA followed by rolling circle amplification (Drmanac et al., 2010).
This generates single strands of coiled head-to-tail DNA called DNA nanoballs (DNBs).
One advantage of rolling circle amplification is that the original copy of DNA acts as the template for each new copy.
This PCR amplification-free process reduces index hopping, which is the misassignment of samples during demultiplexing. This is sometimes a source of error in other sequencing approaches (Costello et al., 2018).
The MGI-indexed BRB-seq or DRUG-seq libraries are now ready for single-strand DNA circularization and DNB preparation with the MGIEasy Circularization kit.
Summary
Overall, the BRB-seq and DRUG-seq library prep workflows are now suitable for use with MGI sequencing technology.
The combined scalability of these ultra-high-throughput 3’ mRNA-seq library prep techniques and MGI sequencers now allows researchers to generate more transcriptomic data quicker, easier, and at lower costs than ever before.
To find out more about our MGI-compatible library prep kits please contact us or visit our BRB-seq and DRUG-seq for MGI product pages.
References
- Alpern, D. et al. (2019) ‘BRB-seq: Ultra-affordable high-throughput transcriptomics enabled by bulk RNA barcoding and sequencing’, Genome Biology, 20(1), pp.1-15. Available at: https://doi.org/10.1186/s13059-019-1671-x.
- Drmanac, R. et al. (2010) ‘Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays’, Science, 327(5961), pp. 78–81. Available at: https://doi.org/10.1126/science.1181498.
- Costello, M. et al.(2018) ‘Characterization and remediation of sample index swaps by non-redundant dual indexing on massively parallel sequencing platforms’, BMC Genomics, 19, 332. Available at: https://doi.org/10.1186/s12864-018-4703-0.




