MERCURIUS™ BRB-seq and MERCURIUS™ DRUG-seq library preparation kits are compatible with MGI sequencing technology. They provide researchers with a scalable, cost-effective bulk 3’ mRNA-seq solution to ultra-high-throughput transcriptomic experiments where MGI sequencers are available.
The bulk 3’ mRNA MERCURIUS™ protocols enable extensive sample multiplexing, which allows researchers to include more biological replicates, experimental conditions, or drug treatments than the standard MGIEasy RNA library prep kit suitable for full-length RNA.
This leads to rapid and robust biological insights that drive drug discovery, fundamental research, and clinical advancements, but there are some important factors to consider with each library prep kit.
Here, we put the different library prep kits head-to-head to see which may be most suitable for your next experiment.
Do you need full-length or bulk 3’ mRNA-seq?
Firstly, researchers must carefully consider the type of RNA-seq method required before deciding between the MGIEasy RNA prep kit or the MERCURIUS™ protocols (Table 1).
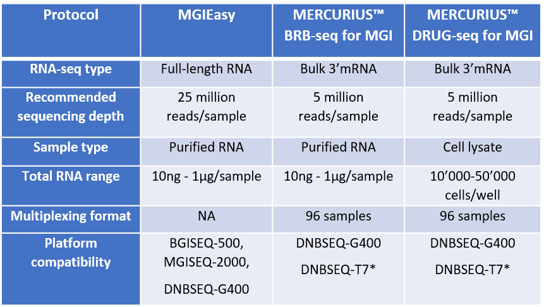
Table 1 – Comparison of MGIEasy RNA prep kits and MERCURIUS™ BRB-seq and MERCURIUS™ DRUG-seq kits for MGI. *Validation in progress.
The MGIEasy RNA prep kit is a full-length RNA-seq approach that provides gene expression information across the whole length of transcripts and also allows researchers to assess alternative splicing and detect novel transcript isoforms (MGI, 2023a).
In contrast, MERCURIUS™ BRB-seq and MERCURIUS™ DRUG-seq are bulk 3’ mRNA techniques that tag the 3’ region of mRNAs with sample-specific optimized barcoded oligo(dT) primers and unique molecular identifiers (Alpern et al., 2019). This enables extensive sample multiplexing and provides researchers with highly accurate read-outs of gene expression from the 3’ end of mRNA transcripts.
A fivefold lower sequencing depth is required to detect the same amount of expressed genes with the bulk 3’ mRNA-seq methods compared to full-length RNA-seq methods, however, information about alternative splicing or transcript isoforms is lost (Table 1). This is an acceptable trade-off for most high-throughput experiments that aim to assess gene expression differences after drug treatment or between conditions.
The choice between full-length and bulk 3’ mRNA library prep methods ultimately depends on the biological question.
Sample prerequisites
The MGIEasy RNA prep kits and MERCURIUS™ BRB-seq library prep kits require RNA to be extracted and purified from samples before starting the workflows (Table 1).
In contrast, DRUG-seq is an RNA-extraction-free protocol where barcoding is performed directly on cell lysates (Table 1). This makes it suitable for ultra-high-throughput experiments such as drug toxicology screens or extensive biobank studies.
For MGIEasy and BRB-seq, each sample requires between 10ng and 1µg of purified RNA. On the other hand, DRUG-seq uses cell numbers as a substitute for RNA amount, with a suggested range of 10,000 to 50,000 cells per well.
In each MERCURIUS™ protocol, the amount of RNA or cells should be uniform across all samples to ensure an even distribution of reads in the library and optimal sequencing results.
Scalability and multiplexing capacity
One of the main advantages of the MGI sequencing technology is that MGI sequencers are the most scalable option for large-scale studies compared to other short and long-read sequencing platforms such as Illumina and Nanopore sequencers.
But, one of the main bottlenecks of MGI and Illumina high-throughput sequencing approaches remains the library preparation step, as each sample usually needs to be handled and prepared individually.
This is the case with the MGIEasy RNA library prep kit as, unlike the MERCURIUS™ protocols, samples cannot be multiplexed in the same tube early on in library preparation.
Without early multiplexing, samples must be processed independently, making the library prep stage time-consuming and expensive due to the excessive amounts of consumables required. Technical errors may also be introduced early in the pipeline, limiting the accuracy of data analysis and the subsequent biological insight gained.
In contrast, with MERCURIUS™ BRB-seq and MERCURIUS™ DRUG-seq, up to 96 samples can be pooled in one tube after sample barcoding at the reverse transcription stage (Table 1). These would therefore be the more appropriate options where a study has large numbers of samples.
Platform compatibility
The most appropriate library preparation method for your experiment also relies on the availability of specific sequencing machines.
For example, MGIEasy RNA library prep is compatible with older sequencing machines such as the BGISEQ-500 and MGISEQ-2000, which the MERCURIUS™ library preps are incompatible with.
If the recent DNBSEQ-G400 machine is available, both MGIEasy and MERCURIUS™ library preps are appropriate.
One significant advantage of the MERCURIUS™ approaches is that they will soon be compatible with the ultra-high-throughput DNBSEQ-T7 sequencing machines, which can produce a staggering 6 Tb of data, and 20 billion sequencing reads from four flow cells (MGI, 2023b). MGIEasy RNA library prep is not yet compatible with this sequencer.
Summary
Thanks to their extensive sample multiplexing and ultra-scalability, the MERCURIUS™ bulk 3’ mRNA-seq library prep kits for MGI sequencing may be a good option for high-throughput transcriptomic studies when researchers have compatible MGI sequencers available and are not focused on alternative splicing or isoform detection.
Conversely, the MGIEasy RNA library prep kit provides information from whole transcripts, so would be more suitable where this is of biological interest.
To learn more about MERCURIUS™ BRB-seq and MERCURIUS™ DRUG-seq, please get in touch with us or visit our product pages.
References
- Alpern, D. et al. (2019) ‘BRB-seq: Ultra-affordable high-throughput transcriptomics enabled by bulk RNA barcoding and sequencing’, Genome Biology, 20(1), pp.1-15. Available at: https://doi.org/10.1186/s13059-019-1671-x.
- MGI (2023a) ‘MGIEasy RNA Library Prep Set’. Available at: https://en.mgi-tech.com/products/reagents_info/id/3
- MGI (2023b) ‘Sequencers’. Available at: https://en.mgi-tech.com/products/




